![]()
Chapter 1
PHYLOGENETIC ANALYSES AND MORPHOLOGICAL INNOVATIONS IN LAND PLANTS
James A. Doyle
Department of Evolution and Ecology, University of California, Davis, CA, USA
Abstract: An increasingly robust phylogenetic framework based on molecular and fossil data clarifies the sequence of evolutionary innovations in land plants. Oogamy and cellular novelties (phragmoplast, plasmodesmata, incipient meristems) evolved in aquatic streptophytes. Invasion of the land entailed interpolation of the sporophyte, jacketed gametangia and sporangia, and air-dispersed spores, followed by stomata. Origin of vascular plants involved branching of the sporophyte and stepwise evolution of vascular tissue. Leaves originated independently in lycophytes and euphyllophytes; in some euphyllophytes leaves were derived from single dichotomous branches, in others from whole branch systems. In seed plants, secondary growth evolved before the seed. Pinnately compound leaves were replaced by simple leaves in coniferophytes. The origin of the angiosperm flower remains unresolved, but bitegmic ovules may be derived from cupules, and the ancestral carpel can be reconstructed as ascidiate. Evolution of double fertilization was a stepwise process that continued within angiosperms; vessels also evolved within the group. Monocots show major reorganization tied to loss of secondary growth, while pentamerous flowers evolved from dimerous within eudicots.
Keywords: evolution; phylogeny; morphology; innovations; land plants; angiosperms.
1.1 Introduction
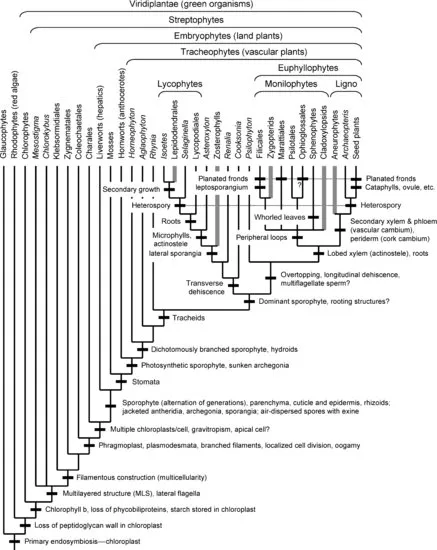
As in other groups of terrestrial organisms, the evolution of land plants involved a series of radiations linked with major evolutionary innovations, many of them clearly adaptations that allowed progressively more efficient and varied occupation of the land environment. The morphology of familiar plants such as the model system Arabidopsis thaliana therefore represents a hierarchical accumulation of structural features that arose at different points on the line from their distant aquatic ancestors, with older advances shared with a successively wider range of relatives. This chapter attempts to summarize the present picture of the sequence of evolutionary innovations in the latest phylogenetic framework, as well as outstanding unresolved issues.
Some of the main events in the evolutionary history of land plants have been recognized since the late nineteenth century. Key insights were recognition of the alternation of haploid and diploid generations, seen in its most basic and obvious form in “bryophytes” such as mosses and “lower vascular plants” such as ferns, and realization that this life cycle persists in modified form into seed plants (Hofmeister 1862; Bower 1890, 1908; Strasburger 1894). However, the details have become much clearer over the past century as a result of many factors, including fossil discoveries that show intermediate stages and character combinations no longer preserved in the living flora, technical advances that revealed new suites of characters at the microscopic and ultrastructural level, development of more explicit methods of analysis of phylogenetic relationships, and the application of these methods to molecular sequence data. Methods of phylogeny reconstruction, many derived from earlier partial insights (notably Zimmermann 1931; Donoghue & Kadereit 1992) but first clearly synthesized in English by Hennig (1966), were elaborated under the rubric of “cladistics” in the 1970s and 1980s and used in analyses of morphological characters. These methods used the principle of parsimony to search for the phylogenetic tree involving the fewest character state changes, on the assumption that this is the tree most consistent with the totality of characters recognized.
Whereas some precladistic discussions assumed that phylogeny could only be approached by consideration of fossils and identification of direct ancestors, which implied that the phylogeny of groups such as angiosperms with a supposedly poor fossil record could not be understood, cladistic methods could be applied to both living and fossil organisms. These methods also made it possible to draw conclusions on the origin of groups and their ancestral states by recognition of closest outgroups without identification of actual direct ancestors. At the level of land plants, many analyses included both fossil and living taxa. There was considerable discussion of the relative importance of the two sorts of data, some arguing that the main relationships among living organisms could be reconstructed without fossil data, which necessarily have far fewer characters due to lack of preservation of parts (Patterson 1981). Others argued that inclusion of fossils was necessary to obtain correct relationships, as in amniote vertebrates (Gauthier et al. 1988), and even when fossils were not required to infer the correct topology of the tree of living organisms, they could be needed to reconstruct the evolutionary steps leading to living clades, which are often separated from their closest relatives by large numbers of morphological changes (Donoghue et al. 1989). In molecular hindsight, morphological cladistic analyses correctly resolved many contentious problems that had plagued earlier intuitive approaches, such as the monophyly of land plants and angiosperms. On other questions, however, such as rooting of the angiosperm phylogenetic tree and relationships among vascular plant and seed plant lines, the results varied from one analysis to another, presumably due to homoplasy (evolutionary convergence and reversal), different interpretations of characters, and variable sampling of both extant and fossil taxa.
This picture has improved dramatically in the past two decades with the accumulation of vast quantities of molecular sequence data from more and more species, which has led to increasingly complete, consistent, and statistically well-supported trees of living organisms. The first analyses of sequences of single genes showed many of the same sorts of inconsistencies seen in morphological analyses. However, as more genes have been sequenced and combined into multigene and even whole-genome analyses, many tentative early results have stabilized and become statistically robust, and with some conspicuous exceptions, most early conflicts between genes have been firmly resolved. These studies started with parsimony analysis, but newer maximum likelihood and Bayesian methods take a more statistical approach to changes on branches. This has led to a role reversal—whereas formerly ideas on the evolution of morphological characters were used to reconstruct phylogenies, phylogenies based on molecular data are now used to reconstruct the evolution of morphological characters, by plotting (optimization) of character states on trees derived from molecular data, using parsimony or likelihood-based methods, thus avoiding dangers of circular reasoning.
For understanding of major evolutionary innovations, a major weakness of the increasingly exclusive reliance on molecular data for phylogeny reconstruction is that purely molecular analyses cannot include fossil taxa, since with the exception of very recent fossils (such as human relatives) no DNA remains. There is much discussion of whether morphological data have any role at all in reconstruction of phylogenies of living organisms (Scotland et al. 2003; Wiens 2004). Because of the vastly greater number of DNA characters, combined analyses of morphological and molecular data tend to be dominated by molecular data. Morphology may still have some role in resolving parts of the phylogeny where molecular analyses of different genes give conflicting or poorly supported results, such as among seed plants and major groups above the base of the angiosperms, but it may be argued that this represents only a brief intermediate phase before all relationships are cemented by genomic data.
Even if molecular analyses completely resolve all relationships among living taxa, other approaches will be needed to address those cases where fossils provide evidence on transitions that are not preserved in the living flora. Molecular analyses consider only relationships among crown groups—where a crown group includes the most recent common ancestor of the living members of a clade and all its derivatives—and not fossils on or attached to the stem lineages leading to these clades, known as stem relatives (Doyle & Donoghue 1993). Integration of these fossils will still require compilation and analysis of morphological data from both the fossils and all relevant living taxa. The ideal approach may be a “total evidence” analysis that combines both molecular and morphological data (Hermsen & Hendricks 2008), but this may not be so easy because of problems in choice of molecular data sets and differences in taxon sampling: single species in molecular data sets, but often higher taxa for which ancestral states have been reconstructed in morphological data sets. A few studies of land plants have used a total evidence approach to integrate morphological data from fossils with molecular data (e.g., Rothwell & Nixon 2006), but in the meantime others have used a “molecular scaffold” approach (Springer et al. 2001; Manos et al. 2007; Doyle & Endress 2010) that integrates fossils by analyzing a morphological data set for both living and fossil taxa with the relationships among living taxa fixed to a “constraint tree” based on molecular data. Such analyses essentially ask what additional insights fossils provide if the relationships inferred from molecular data are correct.
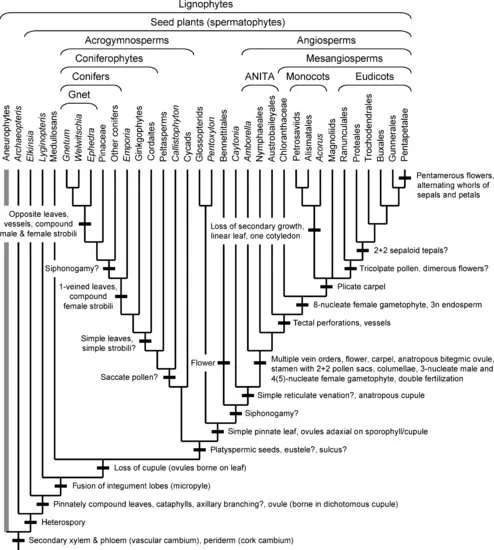
This chapter does not pretend to be a comprehensive review of the literature, but rather a selective though hopefully balanced survey of current ideas and evidence. Currently understood phylogenetic relationships and the placement of morphological innovations are summarized in Figures 1.1 and 1.2. A good general review of land plant phylogeny is provided by Judd et al. (2008). Except where noted, most information on morphological characters of the living and fossil taxa discussed here can be found in standard plant morphology and paleobotany texts (Smith 1955; Gifford & Foster 1989; Stewart & Rothwell 1993; Crum 2001; Taylor et al. 2009) and Graham (1993) for algal outgroups. Not all the innovations mentioned are equally “major” in an evolutionary sense, but some more obscure ones are of interest as providing morphological support for relationships. In most cases, I refer to clades above the ordinal level with anglicized versions of names in the phylogenetic nomenclature of Cantino et al. (2007), and I use quotes to mark traditional paraphyletic groups when these are first mentioned (e.g., “bryophytes”). Hopefully this summary will be useful as a framework for investigations on the developmental-genetic and functional bases of the evolutionary changes inferred.
1.2 Basic innovations in cell structure and life cycle: aquatic streptophytes
Both morphological and molecular phylogenetic analyses confirm the long-standing view that land plants are members of the clade of green organisms, or Viridiplantae, in which other members have been traditionally called green algae. The most conspicuous innovations that unite this clade are the origin of chlorophyll b (in addition to chlorophyll a) and storage of starch in the chloroplasts. Green organisms are in turn linked by molecular data to red algae (rhodophytes) and the unicellular glaucophytes. Molecular phylogenies indicate that these three groups were derived from the line in which the chloroplast first originated by primary endosymbiosis with a cyanobacterium, from which chloroplasts of all other photosynthetic eukaryotes were derived by secondary endosymbiosis (incorporation of a red or green alga) or tertiary endosymbiosis (Delwiche & Palmer 1997; Keeling 2004). The chloroplasts of glaucophytes retain a remnant of the peptidoglycan cell wall seen in free-living cyanobacteria and other eubacteria, whereas both glaucophytes and red algae retain phycobiliproteins, the characteristic photosynthetic accessory pigments of cyanobacteria, which were lost in green plants.
Within green organisms, studies of cell ultrastructure in the 1960s and 1970s led to the view that certain groups of “green algae” are more closely related to land plants than others (Pickett-Heaps 1969, 1972, 1975, 1979; Stewart & Mattox 1975; Graham 1993), such as Charales (complex filaments with whorled branches), Coleochaetales (Chaetosphaeridium, with branched ...