Michael J. McGarvey1 and Michael Houghton2
Summary
There have been major advances recently in several areas of hepatitis C virus (HCV). The array of host receptors important for HCV entry now includes HSGP, LDLR, CD81, SRB1, CLDN1, occludin, and NPC1L1. The interaction of host and nonstructural viral proteins is essential for genome replication. Long-range interactions between cis-acting elements in the 5′ and 3′ UTRs and the NS5B coding region may control the switching between genome replication and genome packaging into virions, as well as modulate translation. Capsid assembly also involves NS5A, NS2, NS3/4A, and viral RNA as well as core protein. HCV assembly is intimately connected to host lipid metabolism and the synthesis of very-low-density lipoproteins (VLDLs) resulting in a lipoviroparticle. Core, NS5A, NS2, NS3, and NS4B can regulate host metabolic processes. HCV proteins have been shown to compromise host defenses at many levels, such as NS3 and NS5A inhibition of interferon alpha and NS5A and NS5B inhibition of apoptosis.
Introduction
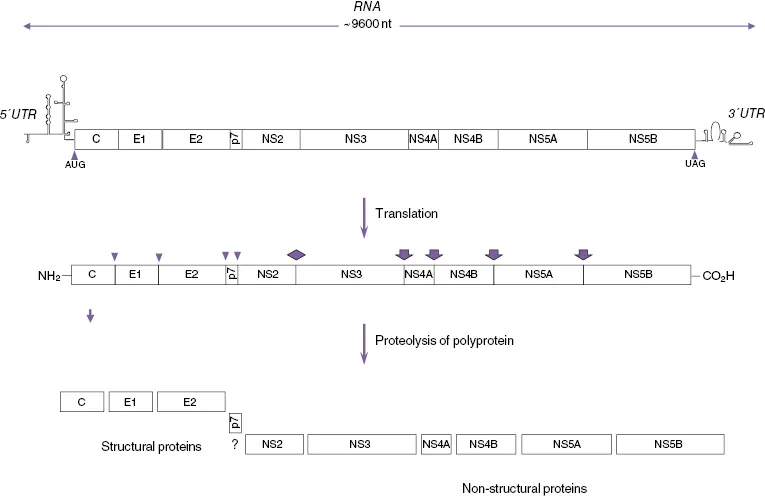
Hepatitis C virus (HCV) is an enveloped positive-strand RNA virus of the genus Hepacivirus in the Flaviviridae family. HCV causes chronic infection in around 80% of cases [1, 2], which then predisposes patients to the sequential development of cirrhosis and primary hepatocellular carcinoma (HCC) [3]. The HCV genome is composed of a positive-stranded RNA molecule of about 9500 nucleotides (nt) [4] containing a single long translational open reading frame (ORF) that encodes a large polypeptide of approximately 3000 amino acids (aa), beginning with the first in-frame methionine codon [5, 6] bounded by 5′ and 3′ untranslated regions (UTRs) of approximately 341 and 230 nucleotides (Figure 16.1). The 5′ UTR of the HCV genome has substantial primary sequence identity with the corresponding region of pestivirus genomes [5, 7], and the nucleotide triphosphatase (NTPase) region of the NS3 helicase has significant sequence identity with those of the pestiviruses and, to a lesser extent, the flaviviruses [5, 8, 9]. Protease and RNA-dependent RNA polymerase (RdRp) sequence motifs, conserved among the pestiviruses and flaviviruses, are also present within the HCV-encoded polyprotein, which along with the more extensively conserved helicase sequence are all similarly collinear among the three types of viral polyproteins [5]. Although these are the only regions of HCV exhibiting significant primary sequence identity with pestiviruses and flaviviruses, the hydropathicity of the HCV-encoded polypeptide is remarkably similar to that of the flaviviruses and, to a lesser extent, to that of the pestiviruses, thus indicating similarities in their basic structures and functions [5]. Nucleotide and protein sequence analyses also show that HCV is more closely related to GBV-B virus than to the other members of the Flaviviridae family, and together they comprise the Hepacivirus genus. It has recently been proposed that the other GBV viruses, GBV-A, GBV-C (HGV), and GBV-D, should be classified as a separate genus, the Pegiviruses [10]. There is substantial sequence variation in the HCV genome resulting from the lack of proofreading of the NS5B RdRp and the subsequent accumulation of errors in replicating HCV genomic RNA. This means that, even in a single infected individual, the HCV genome does not exist as a homogeneous species. Rather, it exists as a quasi-species of closely related but nevertheless heterogeneous genomes [11]. In addition, the process of host selection and adaptation of a rapidly mutating genome has led to the evolution of many distinct HCV genotypes. A large number of studies have been conducted on the phylogeny of HCV. To standardize the various systems that had developed in different laboratories, a uniform system for the nomenclature of HCV genotypes was derived from sequence comparisons in the NS5 region of HCV variants from a worldwide panel. The sequence similarities between members of different genotypes were 55–72%. Similarities between those within related subgroups ranged from 75% to 86%, and individual isolates from each of the clusters showed 88% sequence similarity [12, 13]. Originally six genotypes or clades (1, 2, 3, 4, 5, and 6) with a variable number of subtypes (designated a, b, etc.) for each of these genotypes were identified, and this was used as the standard system for the nomenclature of HCV genotypes [11]. However, more recently a single isolate of a seventh genotype has been identified [14]. In addition, there have been a number of reports of possible recombinant genotypes [15]. Most recently a non-primate hepacivirus with strong homology to HCV has been described [16].
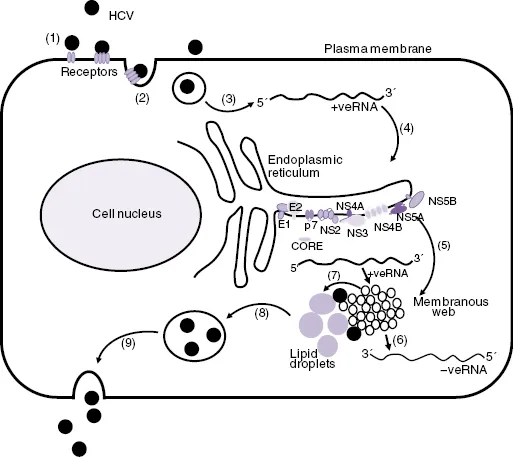
Significant advances in the study of HCV have come from the development of subgenomic replicating HCV RNAs (RNA replicons) as cell-based models for HCV RNA replication. These were developed from full-length HCV genomes in which the sequence coding from core to p7 or from core to NS2 was replaced by a gene cassette that consisted of neomycin phosphotransferase gene (i.e., a “neo”-selectable marker) and a downstream encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). In this construct, the “neogene” is translated under the control of the HCV IRES, and the HCV NS proteins are translated under the control of the EMCV IRES. Human hepatoma 7 (Huh7) cells transfected with HCV replicon RNA are grown in culture medium that contains G418, which only allows the growth of only cell lines that contain the replicating HCV RNA replicons [17, 18]. More recently, the full replication cycle of HCV (Figure 16.2) in cell culture (HCVcc) has been achieved by cloning a HCV genome from a genotype 2a HCV-infected patient in Japan who had fulminant hepatitis (i.e., Japanese fulminant hepatitis-1 [JFH-1]). Full-length RNA was then synthesized from the cloned JFH-1 cDNA, and after transfection into Huh7 cells or the more permissive Huh7.5 derivative cell line, infectious virus particles were produced [19, 20]. These virus particles were then used to infect naïve cells. The JFH-1 HCV replication system and a small number of chimeric viruses derived from JFH-1 and some additional genotypes have become a standard model for HCV replication in laboratories throughout the world [21, 22].
The HCV virion
Particles with a diameter of 45–65 nm have been observed by electron microscopy (EM) in human plasma [23] and in chimpanzee and human liver chronically infected with HCV [24, 25]. Non-enveloped nucleocapsids have also been detected in the plasma of infected chimpanzees [26]. Analysis of the structure of HCV virions from cell culture, using negative-staining electromicroscopy, has shown that they are spherical and pleomophic with a diameter of 40–75 nm. However, using cryoelectron microscopy, it was found that there are two main populations of particles of 60 and 45 nm diameter [27]. The 60 nm particles possess an internal structure that appears to correspond to the capsid and a separate membrane bilayer, and they have a high RNA-to-infectivity ratio and appear to correspond to complete virions. The 45 nm particles lack the membrane bilayer, were less infectious, and may correspond to non-enveloped viral capsids. Analysis of the lipid composition of purified E2-FLAG tagged virus particles by mass spectrometry showed that HCV particles possess a unique lipid composition that is very distinct from all other viruses, with cholesteryl esters accounting for almost one-half of the total HCV lipid content [28]. Electron microscopy showed that the particles were heterogeneous, mostly spherical structures, with an average diameter of about 73 nm, and immuno-EM showed that most E2-containing particles also contained apolipoprotein E (apoE) on their surface [28]. There is evidence that HCV particles are present in the circulation as immune complexes [29] or in association with serum lipoproteins [30–32]. Analysis of virus particles from HCV-infected patients shows that lower density particles have a much higher infectivity than higher density particles [33–35]. Particles of around 100 nm, which are rich in triacylglycerol and probably contain apoB, apoE, and apoC1-C3, have been described [33]. HCV also appears to be associated with apoE to form lipoviroparticles (LVPs), which contain components of very-low-density lipoproteins (VLDLs) [36].
Structural proteins
A large ORF extends throughout most of the HCV RNA genomic sequence and encodes a polypeptide of between 3010 and 3033 amino acids, depending on the source of the viral isolate [5, 37, 38]. As in the case for pestiviruses and flaviviruses, the large HCV ORF encodes a polyprotein precursor that is processed co- and posttranslationally to yield a variety of structural (virion) and nonstructural (NS) proteins (Figure 16.1). Structural proteins are processed from the N-terminal region of the HCV polyprotein precursor, beginning with the 23 kDa basic capsid or core RNA-binding protein (C) followed by two envelope glycoproteins – E1 (33–35 kDa) and E2 (68–72 kDa) (Figure 16.2).
Core protein and virus assembly
The expression of the HCV structural protein-coding region, by cell-free translation in the presence of microsomal membranes or in tissue culture cells, produces a 23 kDa core protein of 191 amino acids. This indicates that the cleavage of the core protein from the polyprotein is dependent on signal peptidases in the endoplasmic reticulum (ER) [29, 39, 40]. HCV core protein has been identified in the sera of HCV-infected humans as a 21 kDa protein. The core protein binds to membranes in vitro and in transfected cells, and has been shown to be associated with the cytoplasmic side of the ER. E1 was precipitated by anticore antibodies in the presence of core protein, showing that core interacts with E1. In contrast, the E2 protein is not co-precipitated with anticore antibodies in co-expression experiments. Following signalase cleavage, the E1 signal sequence may act, at least in part, to retain the upstream C protein in the ER in an analogous manner to that of the flaviviruses [41, 42]. Little E1 is exposed on the cytosolic side of the ER; therefore, the interaction between core and E1 probably takes place in the ER membrane, and deletion studies indicate that the C-terminal regions of both proteins are important for their interaction [43]. Both membrane-bound and membrane-free core exist in dimeric and multimeric forms. Core proteins, which had the hydrophobic C-terminal domain deleted, also translocated to the nucleus. One of the three clusters of basic amino acids in the core protein, PRRGPR (aa 38–43), is similar to the nuclear localization signal found in human papilloma and human herpe...