
Single-Cell Omics
Volume 1: Technological Advances and Applications
- 490 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Single-Cell Omics
Volume 1: Technological Advances and Applications
About this book
Single-Cell Omics: Volume 1: Technological Advances and Applications provides the latest technological developments and applications of single-cell technologies in the field of biomedicine. In the current era of precision medicine, the single-cell omics technology is highly promising due to its potential in diagnosis, prognosis and therapeutics. Sections in the book cover single-cell omics research and applications, diverse technologies applied in the topic, such as pangenomics, metabolomics, and multi-omics of single cells, data analysis, and several applications of single-cell omics within the biomedical field, for example in cancer, metabolic and neuro diseases, immunology, pharmacogenomics, personalized medicine and reproductive health.This book is a valuable source for bioinformaticians, molecular diagnostic researchers, clinicians and members of the biomedical field who are interested in understanding more about single-cell omics and its potential for research and diagnosis.- Covers not only the technological aspects, but also the diverse applications of single cell omics in the biomedical field- Summarizes the latest progress in single cell omics and discusses potential future developments for research and diagnosis- Written by experts across the world, bringing different points-of-view and case studies to give a comprehensive overview on the topic
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
An Overview of Single-Cell Isolation Techniques
† Department of Paediatrics, Schulich School of Medicine and Dentistry, Western University, London, ON, Canada
‡ School of Life Sciences, Tsinghua University, Beijing, China
Abstract
Keywords
Acknowledgments
6.1 Introduction
6.2 Methods of Single-Cell Isolation
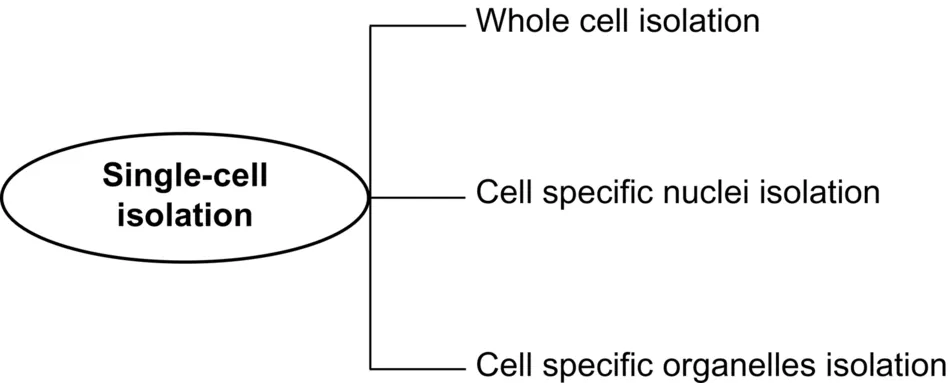

6.2.1 Manual Micromanipulation

Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Contributors
- About the Editors
- Preface
- Section I: Overview of Single-Cell Omics
- Section II: Omics Technologies in Single-Cell
- Section III: Data Analysis in Single-Cell Omics
- Index