![]()
Section V
Fermentation Microbiology and Biotechnology
Tools, Monitoring, and Control of Fermentation Processes
![]()
14Functional Genomics
Current Trends, Tools, and Future Prospects in the Fermentation and Pharmaceutical Industries
Surendra K. Chikara , Toral Joshi , and Mahmoud M. A. Moustafa
CONTENTS
14.1Introduction
14.2Microarrays: Role in Functional Genomics
14.2.1Applications of Microarray to the Study of Genome Structure
14.2.2Applications of Microarray to the Study of Gene Expression and Profiling
14.3Microarray Quality Control
14.3.1Affymetrix
14.3.2NimbleGen MS 200 Microarray and Scanner
14.3.3Agilent DNA Microarray and Scanner
14.4Microarray Applications
14.4.1Gene Expression Profiling
14.4.2CGH
14.4.3Detection of SNP
14.4.4Chromatin Immunoprecipitation
14.4.5Transcriptome Analysis
14.4.6Application of DNA Microarray in the Pharmaceutical Industries
14.4.7Microarray Future Prospects
14.5Next-Generation DNA Sequencing
14.5.1First-Generation Sequencers
14.5.1.1Sanger’ s Sequencing Method
14.5.2Second-Generation Sequencers
14.5.2.1Roche GS FLX (Pyrosequencing)
14.5.2.2Illumina Genome Analyzer
14.5.2.3Applied Biosystems SOLiD™ 4 System
14.5.2.4Ion Personal Genome Machine Sequencer
14.5.3Third-Generation Sequencers
14.5.3.1Helicos
14.5.3.2Pacific Biosciences
14.5.3.3VisiGen
14.5.3.4BioNanomatrix
14.6Comparison of the Sequencing Techniques
14.7Discovery of SNP
14.8Transcriptome Analysis
14.8.1Gene Expression: Sequencing the Transcriptome
14.8.2Applications of Transcriptomics Analysis
14.8.3Small RNA Analysis
14.8.3.1Techniques for RNA Analysis
14.8.3.2Discovering NC-RNA
14.9Epigenetics
14.10Chromatin Immunoprecipitation, Chip-Seq Technique
14.11Metagenomics
14.12Genome Analysis
14.12.1Whole-Genome Sequencing
14.12.2De Novo Assembly
14.12.3Annotation of Genome Assembly
14.12.4Genome Mapping of Next Generation Sequencing Data
14.12.5Targeting Resequencing
References
“ When you are face to face with a difficulty, you are up against a discovery.”
Lord Kelvin
14.1Introduction
Functional genomics is a discipline and enterprise that exploits the vast wealth of data produced by genome sequencing projects. A key feature of functional genomics is their genome-wide approach, which invariably utilizes high-throughput technologies and relies on sophisticated analytical tools.
Understanding the genome sequence and its relevance for various applications is therefore central to the development and production of new biopharmaceuticals. Current advances in bioinformatics and high-throughput technologies such as microarray analysis have revolutionized our perception and understanding of the molecular mechanisms underlying normal and abnormal (dysfunctional) biological functions. Microarray studies and other genomic techniques are also inspiring the discovery of new targets for disease treatment and control, thus aiding drug development immunotherapeutics, and gene therapy (Zhao et al., 2011; Beitelshees et al., 2017). Current trends and challenges in the field include
• Exploitation of new innovations in nanotechnology and microfluidics for the development of low-cost technologies for sequencing and genotyping as well as for the identification and confirmation of functional elements that do not encode protein (e.g., introns, promoters, telomeres , and regulatory and structural features);
• Telomere : A telomere is a region of repetitive DNA located at the end of chromosomes to protect them from degradation.
• In vivo , real-time monitoring of gene expression and functional modification of gene products in all relevant cell types using large-scale mutagenesis, small-molecule inhibitors, and knockdown approaches;
• Identifying genes and pathways that have thus far proved difficult to study biochemically and deciphering cellular phenomena related to the networking of metabolic pathways;
• Monitoring of membrane proteins, modified proteins, and regulatory proteins of low concentrations; and
• Correlation of genetic variation to human health and disease using haplotype and comprehensive variation information to provide large databases that are amenable to statistical methods.
The challenges faced in the human genome project and the need for high-throughput capacity have led to the development of next-generation sequencing technology (NGST), which has dramatically accelerated biological and biomedical research because it renders the comprehensive analysis of genomes, transcriptomes, and interactomes inexpensive, which should be helpful toward achieving the goal of personalized medicine and designer crops (Horner et al., 2010). Of pharmaceutical relevance is sequencing of multiple strains of pathogens to monitor drug resistance and pathogenicity. Resequencing of selected regions to search for human variation in population and for tumor profiling to guide cancer therapies is also of great interest. In this chapter, we review current trends and methodologies in functional genomics and assess their relevance, strengths, and weaknesses.
Completion of the human genome project signaled a new beginning for modern biology, one in which most biological and biomedical research will be conducted in a sequence-based fashion. It is now possible to deconstruct the genomic sequences of multiple organisms with the view to unravel the interrelationship between the physiology or the pathogenicity of a given organism and its genome constituent parts (genes) (Cook Degan, 1991; 56Johnson, 1992; Greenhalgh, 2005; Evans, 2010). The deconstruction of the genome to assign biological functions to genes, groups of genes, and particular gene-gene or gene-protein interactions is well underway and documented in MEDLINE (Patterson and Gabriel, 2009). These functions may be directly or indirectly the result of a gene’ s transcription (Mamanova et al., 2010). A computationally intensive branch of functional genomics has emerged as a result of the practical implementation of technologies to assess gene expression of thousands of genes at a time. The ability to comprehensively measure gene expression affords an excellent opportunity to further expand a target-oriented approach as opposed to the conventional hypothetical-based approach.
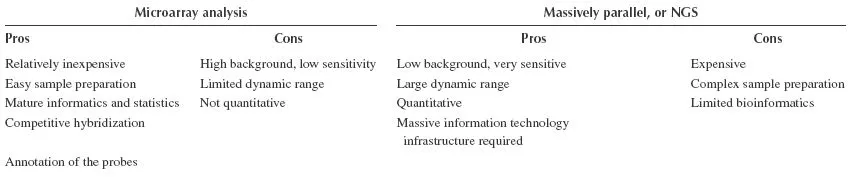
The need to generate, analyze, and integrate large and complex sets of molecular data has led to the development of whole-genome approaches, such as microarray technology, to expedite the process of translating molecular data into biologically meaningful information (Miyake and Matsumoto, 2005). The comparison of the strengths and weakness of next-generation sequencing and microarray techniques is illustrated in Table 14.1 (Asmann et al., 2008).
TABLE 14.1 The Strength and Weaknesses of NGS and Microarray Techniques
Source: I...