![]()
Chapter 1
The Sampling and Sample Preparation Problem in Microbial Metabolomics
Walter M. van Gulik, André B. Canelas, Reza M. Seifar, and Joseph J. Heijnen
1.1 Introduction
The reproducibility and accuracy of analysis methods has expanded rapidly over the past years. Hyphenated methods, for example, GC-MS and LC-MS/MS (liquid chromatography-mass spectrometry), are now almost routinely used for (semi-) quantitative analysis of tens to hundreds of metabolites in biological samples. It should be realized, however, that the quality of the applied analytical techniques alone is not sufficient to obtain meaningful results. Other important aspects, which are often overlooked, are the methods that are applied to withdraw the samples from a microbial cultivation and the subsequent sample processing procedures. Immediately after withdrawal of the samples, the metabolism should be arrested to preserve the metabolic snapshot represented by the sample by preventing further metabolic conversions. Subsequently, an appropriate extraction method should be applied, which guarantees complete and unbiased release of all metabolites from the cells without significant degradation and inactivation of enzymatic activity. Finally, an accurate and high throughput analytical platform is required for metabolite quantification. In this chapter, we focus on the essential requirements of the sampling and sample processing procedures as well as the methods to validate if these requirements are met in practice.
1.2 Microorganisms and Their Properties
Microorganisms show very large differences in properties, for example, size, structure, degree of complexity, heterogeneity, physiology, nutrient requirements, and so on. These aspects should be considered beforehand because they can be important for the choice of the sampling and sample processing methods to be used. They also determine the meaning of the obtained results, for example, the interpretation of metabolite measurements in cell extracts of prokaryotic microorganisms, which contain only one compartment (cytosol), is more straightforward compared to measurements in cell extracts of eukaryotes. As eukaryotic (micro)organisms contain several compartments (cytosol, mitochondria, peroxisomes, etc.) in which the concentrations of metabolites can be very different, the interpretation of the measurements, which are in fact whole cell averages, can be problematic if the metabolites quantified are present in more than one cellular compartment.
1.3 Sampling Methods
1.3.1 The Need for Rapid Sampling
As quantitative metabolomics aims at obtaining accurate snapshots of intracellular metabolite levels, the withdrawal of samples from the microbial culture under study should be sufficiently fast to prevent significant changes in metabolite levels arising from ongoing metabolic activity. This is particularly important if samples are taken from cultures for which one of the medium components is the growth-limiting nutrient (e.g., chemostat or fed-batch cultivations). If the sampling procedure requires too much time, the small amount of growth-limiting nutrient present in the sample will be rapidly exhausted, which will result in changes in metabolic fluxes and consequently in changes in intracellular metabolite levels. Similarly, if samples are taken from aerobic cultivations, the amount of oxygen dissolved in the liquid phase will be rapidly exhausted because of the consumption of oxygen by the cells combined with the very limited solubility of oxygen in water.
It should be realized that the speed at which the ongoing metabolic activity of the cells in the culture sample occurs, and thus the time of exhaustion of the limiting nutrient and/or the available oxygen in the liquid phase, directly depends on the biomass density and the growth rate of the culture from which the sample is withdrawn. For relatively high density and fast growing cultures, this could happen in a few seconds. Considering the pool sizes of intracellular metabolites, which range from 10−3 to 102 μmol g−1 dry cell mass [1–3] and the fluxes through the metabolic network, the apparent turnover times of metabolite pools range from fractions of a second to tens of minutes [4]. This implies that to obtain meaningful results a dedicated rapid sampling system is required, whereby the residence time of the sampled culture broth in the sampling system is in the subsecond range.
Furthermore, the metabolic activity of the cells should be arrested as soon as the sample enters the sampling vial. This has generally been accomplished by introducing the culture broth with sufficient velocity (i.e., as a liquid jet) into a dedicated quenching solution (Section 1.4) such that it is instantaneously mixed. A common approach is to use a cold aqueous solvent mixture, for example, 60% aqueous methanol, to cool down the sample below −20 °C, thereby minimizing the enzymatic activity. It has been shown that with this method effective quenching of metabolism is obtained only if the mixing of the sample with the cold aqueous quenching liquid is instantaneous [5]. Another approach is to combine quenching with extraction, by sampling directly into the extraction solution (e.g., perchloric acid) or by integrating extraction in the sampling mechanism itself [6].
1.3.2 Sampling Systems
An overview of different rapid sampling systems, both manually operated and fully automated, has been provided by Schaedel and Lara [7]. Apart from being sufficiently fast, other important requirements for rapid sampling systems are that the amount of sample withdrawn is reproducible; the dead volume is negligible (to prevent that stagnant liquid residing in the system is mixed up with the sample); and especially for sampling during highly dynamic experiments, the time between subsequent samples is sufficiently small to fully capture the dynamics.
A convenient manually operated rapid sampling system for withdrawal of broth samples from bench scale bioreactors has been described by Lange et al. [8]. This system consists of a sampling port, inserted in the wall of the bioreactor, with an internal diameter of 1 mm, which is connected to a tube adapter. Sampling is started by removing the dead volume by flushing into the waste. Subsequently, the sample tube is evacuated and directly thereafter, the sample is withdrawn from the bioreactor, facilitated by the vacuum in the sample vial and a slight overpressure in the bioreactor. The liquid flows and evacuation of the sample tube are controlled by electromagnetic pinch valves operated by a timer, allowing the sample volume to be precisely adjusted, that is, with a standard deviation of less than 2%. The authors reported that with this system, they could withdraw samples of 1 ml from a bioreactor, operated at an overpressure of 0.3 bar, within 0.7 s. The residence time of the sample in the system was below 100 ms. However, with this system, the sampling frequency could not be increased much above 1 sample per 5 s because of the many manual handlings that had to be performed. Therefore Schaefer et al. [9] developed a completely automated sampling device, whereby the sampling vials were fixed in transport racks, which were moved by a step engine underneath a continuous jet of culture broth, with a flow rate of 3.3 ml s−1, from a stirred tank bioreactor. In this way, each sampling vial, which contained a cold quenching solution, could be filled within 220 ms resulting in a sampling rate of approximately 4.5 samples per second. This automated sampling device was applied to investigate the intracellular metabolite dynamics of glycolysis in Escherichia coli after rapid glucose addition to a glucose-limited steady state culture. A disadvantage of this approach is the large amount of broth that is withdrawn, requiring a relatively large bioreactor.
A different approach to integrated sampling, quenching, and extraction from a bioreactor culture has been published by Schaub et al. [6]. Hereby, short time heating of the sample is used as the procedure to quench all metabolic activity, at the same time extracting the metabolites from the cells. This is achieved by using a helical coil heat exchanger that allowed continuous withdrawal of the sample from a bioreactor, whereby the broth was rapidly heated to 95 °C. The helical geometry was chosen to enhance radial mixing, thus improving the plug-flow characteristics of the system. After extraction, the cell debris can be removed by filtration. This sampling device allows withdrawing approximately 5 samples per second. The method has been applied to the analysis of the growth-rate-dependent in vivo dynamics of glycolysis in E. coli [10].
1.4 Quenching
1.4.1 Quenching Procedures and Their Properties
Different sampling techniques can be combined with various quenching procedures. An essential property of a quenching procedure is that it should accomplish that all metabolic activity is arrested in the moment of sampling. This can be achieved in various ways, for example, by injecting the sample in a solution with an extreme pH (highly acidic or alkaline), by fast heating and subsequent cooling of the sample (see above), by fast cooling to a temperature below −20 °C or by combining different approaches. Rapid cooling can be achieved by injecting the sample in liquid nitrogen or into a cold quenching solution. Examples are cold perchloric acid ( − 20 °C), 1 M alcoholic KOH 50% (v/v) methanol at −20 °C or cold aqueous 60% (v/v) methanol at −40 °C.
An important aspect in deciding which quenching method should be used for a particular case is whether the complete broth sample could be extracted (i.e., cells with the surrounding medium) or whether the cells should be separated from the surrounding medium before extraction. Hereby, it should be realized that in common laboratory-scale bioreactor cultivations, the volume of the surrounding medium is 2 orders of magnitude larger than the total cell volume.
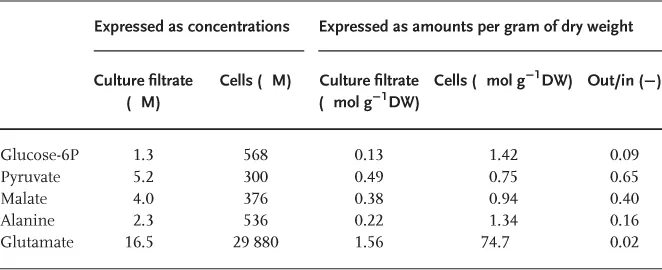
This implies that even low concentrations of metabolites outside the cells could compromise the quantification of the intracellular metabolite levels, if the metabolite measurements are carried out in extracts from total broth samples. This is illustrated by the results of metabolite measurements carried out in glucose-limited chemostats of E. coli, of which the results are shown in Table 1.1. As shown in Table 1.1, the measured metabolite concentrations in the culture filtrate are 2–3 orders of magnitude lower than the intracellular concentrations. Nevertheless, if the intracellular and extracellular levels are expressed as amounts per gram biomass dry weight (DW) present, it appears that for some metabolites (in this example malate and pyruvate) the amount present in the extracellular medium can be quite significant (around 50%). Thus, if metabolite measurements are carried out in extracts of total broth samples, the levels of certain metabolites will be significantly overestimated.
Table 1.1 Example of Extracellular and Intracellular Metabolite Levels Measured in a Glucose-Limited Chemostat of Escherichia Coli K12 MG1655 [3]. To Obtain Intracellular Concentrations a Cellular Volume of 2.5 ml g−1 of Dry Weight was Used
In order to separate the cells from the surrounding medium before the metabolite extraction step, a quenchin...