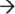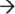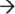![]()
John Wiley & Sons, Inc.
How to cite this article:
WIREs RNA 2012, 3:443–454.
© 2012 John Wiley & Sons, Ltd.
DOI: 10.1002/wrna.1110
Focus Article
RNA decay: a novel therapeutic target in bacteria
Tess M. Eidem,1 Christelle M. Roux2 and Paul M. Dunman1,2*
1Department of Microbiology and Pathology, University of Nebraska Medical Center, Omaha, NE, USA
2Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, NY, USA
The need for novel antibiotics is greater now than perhaps any time since the pre-antibiotic era. Indeed, the recent collapse of most pharmaceutical antibacterial groups, combined with the emergence of hypervirulent and pan-antibiotic-resistant bacteria have, in effect, created a ‘perfect storm’ that has severely compromised infection treatment options and led to dramatic increases in the incidence and severity of bacterial infections. To put simply, it is imperative that we develop new classes of antibiotics for the therapeutic intervention of bacterial infections. In that regard, RNA degradation is an essential biological process that has not been exploited for antibiotic development. Herein we discuss the factors that govern bacterial RNA degradation, highlight members of this machinery that represent attractive antimicrobial drug development targets and describe the use of high-throughput screening as a means of developing antimicrobials that target these enzymes. Such agents would represent first-in-class antibiotics that would be less apt to inactivation by currently encountered enzymatic antibiotic-resistance determinants. © 2012 John Wiley & Sons, Ltd.
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Infectious diseases are the second-leading cause of death worldwide.1 Despite this, there has been a mass exodus of pharmaceutical antimicrobial discovery programs, leaving a void in the drug pipeline that, without intervention, will inevitably result in a healthcare crisis. Indeed, the Infectious Diseases Society of America recently warned of antibiotic-resistant ESKAPE bacterial pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) and the desperate need for new agents to treat these insidious organisms.2 Most current antibiotics are derivatives of molecules discovered over 50 years ago and are losing their foothold as effective means of treating infections due to the emergence of antibiotic resistance.3 Simply put, bacterial antibiotic resistance is outpacing new drug development making it imperative to expand antibiotic drug development to other essential cellular processes in order to create novel agents for the therapeutic intervention of current and emerging antibiotic-resistant bacteria.4 RNA turnover is one such essential biological process that is rich in antimicrobial targets but has not been exploited for antibiotic drug discovery. Accordingly, this review is intended to bring to light the fundamental differences in RNA turnover between host and bacterial pathogen, distinguish those ribonucleases (RNases) that are attractive antibacterial targets, and provide methods to take advantage of these targets for drug development with the ultimate goal of expanding our antibiotic arsenal.
mRNA TURNOVER: PATHOGEN AND HOST
Many currently available antibiotics target essential pathways involved in cell wall synthesis, folate metabolism, protein translation, RNA transcription, or DNA replication.3 These antibiotics are engineered to exert broad antimicrobial activity against an array of bacterial pathogens by targeting essential prokaryotic enzymes within the aforementioned pathways without causing off-target toxic effects toward human counterparts. In that regard, a simple comparison of the physiological characteristics of messenger RNA (mRNA) illustrates wholesale differences between the host and pathogen. For instance, bacteria couple transcription and translation and their mRNA is degraded rapidly (average half-life of ≤2.0 min), does not bear a 5′ 7-methylguanosine (m7G) cap, and is rarely 3′ polyadenylated. Mammalian cells diverge from their prokaryotic ancestors in that they compartmentalize their RNA metabolic steps and their mRNA has a longer half life (minutes to days), is 5′ m7G capped, and is polyadenylated at the 3′ terminus.5 Therefore, it is not surprising that the molecular machinery that governs bacterial and eukaryotic mRNA degradation differs, and consequently these differences could be exploited for antibiotic drug discovery. As a prerequisite to this approach, one must first appreciate the basic similarities and differences in transcript turnover between the host and pathogen, the RNases involved, and their properties, which are briefly described below. For a more comprehensive report of RNA degradation in these two kingdoms, please refer to several recent excellent reviews.6,7,8,9,10,11
The major initiator of bacterial mRNA decay is considered to be a multi-protein complex termed the RNA degradosome. This complex is best characterized in the Gram-negative model organism,
Escherichia coli, and consists of at least four subunits: RNA helicase B (RhlB), enolase, polynucleotide phosphorylase (PNPase), and RNase E (
Figure 1(a)).
12 RNase E is the central component of the
E. coli degradosome, establishing a scaffold for the assembly of other degradosome subunits and performing the initial endoribonucleolytic event during substrate mRNA decay.
12 RNase E preferentially cleaves 5′ monophosphorylated transcripts, thus the rate of mRNA decay is accelerated by the enzyme RppH, which converts the 5′ triphosphate group to 5′ monophosphate.
13 Resulting cleavage products are subsequently digested in a 3′
5′ fashion by the concerted activities of the degradosome-associated exoribonuclease PNPase and RhlB RNA helicase or by degradosome-independent 3′
5′ exoribonucleases, such as RNase II and RNase R.
12,13,14,15,16, Other endoribonucleases also contribute to mRNA degradation, including RNase G, RNase I, and RNase III.
17,18,19 Most of these RNases cannot degrade to the single nucleotide, resulting in short RNA fragments that are further broken down by the enzyme Oligoribonuclease (Orn; 3′
5′ exoribonuclease).
20 Additionally, the endoribonuclease RNase P is known to cleave mRNA transcripts that contain riboswitches and can cleave near stem-loop structures within
E. coli mRNAs.
21,22,23 Resulting cleavage products contain a 5′ loop structure that acts to stabilize select transcripts.
23 RNase E, Orn, and RNase P are essential enzymes in the Gram-negative model organism
E. coli, thus they may be good antibiotic drug discovery targets.
24,25,26