
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Nanomedicine for Drug Delivery and Therapeutics
About this book
This book describes a broad area of nanomedicine which involves mainly applications, diseases, and diagnostics. The comprehensive coverage provides researchers, academics, and health specialists with a great tool, that includes techniques applicable to various uses.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Nanomedicine for Drug Delivery and Therapeutics by Ajay Kumar Mishra in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biotechnology. We have over one million books available in our catalogue for you to explore.
Information
Part 1
NANOMEDICINE
Chapter 1
High-technology Therapy Using Biomolecules or Synthetic Compounds for HIV Inhibition
Abstract
The shortcomings of current treatment of AIDS range from undesirable side effects, incomplete eradication of human immunodeficiency virus (HIV) and an increase in the emergence of drug resistant viral strains. Owing to these limitations, there has been a paradigm shift in the approach of researchers as they now focus on the development of new drugs. More convenient drugs will have enhanced activity, lesser or no side effects and satisfactory delivery potential. Various approaches under investigation use biomolecules and metals and/or synthetic compounds with the potential to inhibit viruses or affect their binding sites on the host cells. Techniques such as gene therapy or metal-based therapy emerge from this concept and have so far contributed promising results for the control of the HIV virus. This chapter explores current developments in gene and metal-based therapies (enhanced by nanotechnology), with respect to the design of effective drugs for the treatment of HIV infection.
Keywords: Antisense oligonucleotides, chimeric oligonucleotides, ribozymes, RNA interference, metal complex, metallodrug, nanoparticles, HIV, therapy
1.1 Gene Therapy Including RNA High-Technology Against HIV
1.1.1 Introduction
Recent efforts in scientific research have allowed the development of a new approach in the fight against HIV-1, called gene therapy. It is a process by which new genetic information is introduced into patients’ cells with a resulting therapeutic benefit, potentially applicable for the treatment of HIV infection. The principle of this new technique resides in the silencing or knocks out of gene expression at the mRNA level [1]. There are various molecules used to generate the loss of cell’s or organism’s functions: Antisense sequences, chimeric oligonuceotides, ribozymes and the small interfering RNA (siRNA) [1]. The main steps involved in an anti-viral gene therapy strategy include:
- Selecting the target for intervention (viral or host function).
- Designing, constructing and expressing the inhibitory gene (RNA decoys, transdominant negative gene product, catalytic RNA, others).
- Selecting the vehicle for gene delivery: defective viral vectors (retroviral [retv], HIV, adenoassociated virus [AAV], others]; liposomes; receptor-ligand mediated, and; other.
- Selecting the mode of intervention: ex vivo modification and manipulation of target cells or direct injection of genetic information (“naked” DNA) into accessible tissue for augmenting immune responses.
1.1.2 Antisense Sequences Technology
In molecular biology, the strand of the gene that carries the information is called the sense strand and the strand complementary to the former is called antisense. What happen normally in plant and animal cells is that the DNA sense strand is transcribed to a messenger RNA (mRNA) in the nucleus, and then the mRNA is transferred in the cytoplasm and translated into protein which can be enzyme or structural protein.
For decades, biologists have realised that they can interfere with this process to modify or inhibit the expression of the genetical information on the DNA or RNA, allowing them to determine the function of specific gene or designing a therapeutic method. One of the first works was targeted at the inhibition of viral growth, using antisense oligonucleotides (tridecamer oligonucleotide) as a hybridization competitor to inhibit Rous sarcoma virus replication [2, 3]. In principle, this technique relies on the use of a sequence, complementary to a specific mRNA that can inhibit its expression and then induce blockade in the transfer of genetic information from DNA to protein or following hybridization, the two strands can form mini double helices that can be recognized or not by the RNase H [4].
Antisense oligonucleotide usually consist of 15–20 nucleotides which are complementary to their target mRNA. The design of appropriate antisense oligonucleotide has to consider the resistance to degradation by the intracellular endonucleases and of course accessible sites on targeted mRNA which are not similar to other genes. Information about the structure of target gene could be obtained by use of RNase H mapping allowing annealing reactions with arrays of antisense species [5, 6].
On the basis of the mechanism of action, two classes of antisense oligonucleotides can be discerned:
- The RNase H-dependent oligonucleotides, which induces the degradation of mRNA.
- The steric-blocker oligonucleotides, which physically prevent or inhibit the progression of splicing or the translational machinery (Figure 1.1).
Figure 1.1 Oligonucleotide act as steric block and prevent binding of important regulatory proteins [7].
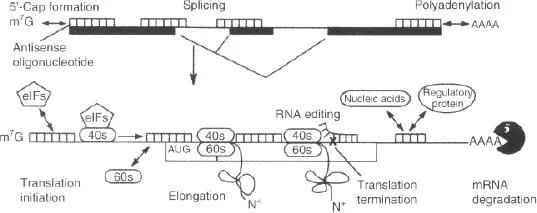
1.1.2.1 RNase H-dependent Mechanism
RNase H is a ubiquitous enzyme that hydrolyses the RNA strand of an RNA/DNA duplex [8]. This enzyme can be activated by some antisense oligonucleotides among which the widely used phosphorothioate [9, 4]. The RNase H-dependent mechanism is quite specific and can induce at 80–90% the inhibition of protein expression from targeted gene: As an example of action of RNase H in retroviruses, Telesnitsky and colleagues [10] explained that during reverse transcription, the viral RNA serves as template for polymerization of minus-strand DNA. RNase H-mediated cleavage of the viral RNA is necessary to free the minus strand for plus-strand DNA synthesis. Studying an RNase H mediated retrovirus destruction, Matzen et al. [11] reported that it resulted from double cleavage of the double stranded DNA (from RNA transcription) at the polypurine tract-U3 junction releasing a 3′ end of the polypurine tract RNA that serves as primer for strand synthesis, and a second cut at the polypurine tract-U3 junction facilitates removal of the primer. In fact, an antisense oligonucleotide complementary to the polypurine tract creates an RNA-DNA duplex that mimics the structure recognized by the reverse transcriptase, leading to premature cleavage of viral RNA at the polypurine tract-U3 junction before reverse transcription.
RNase H dependent oligonucleotide has the advantage that, unlike steric blocker oligonucleotides, which are efficient when binding only at 5′-AUG initiating codon region, phosphorothioate for example can inhibit protein expression when targeted to widely separated areas in the coding region [12, 13, 14]. Deriving from the replacement of a non-bridging oxygen with sulphur on the first chemically synthesized modified oligonucleotides (methylphosphonates); phosphorothioates are the most widely studied oligonucleotides because of their nuclease stability and relative ease of synthesis, their frequent use as antisense effector molecules, result among other from the fact that they are capable of activating RNase H activity [4]. Phosphorothioates were first used as antisense oligonucleotides for the inhibition of HIV replication by Matsukura and coworkers [15]. However phosphorothioates have also been found to have nonspecific interaction, triggering non-antisense effects and stimulating irrelevant cleavage [14], leading to the development of new antisense with more specific action.
For most antisense approaches, target RNA cleavage by RNase H is desired in order to increase antisense potency. Hence in the second generation of antisense oligonucleotides, gapmers were develop and consist of a central stretch of DNA or phosphorothioate DNA monomers and modified nucleotides such as 2′-O-methyl RNA at each end. It was reported that the end blocks prevent nucleolytic degradation of the AS-ON and the contiguous stretch of at least four or five deoxy residues between flanking 2-O-methyl nucleotides enabling activation of Escherichia coli and human RNase H, respectively [16]. Further works to improve RNase H dependent oligonucleotides has led to the development of 2′-deoxy-2′-fluorob-D-aribino nucleic acid (FANA) which was the first uniformly sugar-modified antisense oligonucleotide reported to induce RNase H cleavage of a bound RNA molecule [17] and cyclohexene acids (CeNA) which are characterized by a high degree of conformational rigidity of the oligomers and are resistant to nucleolytic degradation.
Few authors reported on the ease of RNase H dependent oligonucleotides for the inhibition of HIV: Lederman et al. [18] inhibited HIV-1 infection by enhancing the binding of phosphorothioate to the V3 loop of HIV-1 gp120. An in vitro experiment conducted by Veal and Byrn [19] allowed them to successfully demonstrate RNase H cleavage of HIV-1 mRNA mediated by phosphorothioate antisense oligonucleotides complementary to the gag region of the HIV-1 genome.
1.1.2.2 Steric-blocker Mechanism (RNase H-independent Mechanism)
The initial concept of gene therapy was based on the formation of an RNA-DNA duplex that sterically blocked the RNA, resulting in inhibition of gene expression and consequently of viral replication [20]. Further development of this approach has led to precise elaboration of the action of steric-blockers as inhibitor of mRNA translation initiation as well as RNA processing (they can inhibit intron excision, a key step in the processing of mRNA). Splicing occurs during the maturation step and can be inhibited by the hybridization of an oligonucleotide to the 5′ and 3′ regions involved in this process [21]. Such inhibition can lead to the lack of expression of a mature protein [22, 23] or to the correction of aberrant and the restoration of a functional protein [24, 25]. Inhibition of RNA translation by second generation oligonucleotides is mainly attributable to the disruption of the ribosomes or by physically blocking the initiation [26] or elongation steps of protein translation. However the effective target region of a steric block oligonucleotide for inhibiting translation is mainly limited to the 5′-UTR and the start codon region of mRNA, therefore reducing it possible use in antisense therapy. Despite its limitation, steric block remain a viable approach, because of its ability to specifically modulating gene expression, thus lowering off-target effects, compared to conventional antisense. Some of the steric blockers are 2′-O-methyl and 2′-O-methoxy-ethyl RNA oligonucleotides (second generation), peptide nucleic acids (PNAs), N3′-P5′ phosphoramides (NPs) and locked nucleic acid (LNA) (third generation) which effects on the gene expression vary from one to another, but in general they initiate (as above) a blockade of the transcription or the translation by either preventing RNA polymerase action or by hindering the maturation of mRNA for translation [21]. The process of the N3′-P5′ PN oligonucleotides is not well known, and some authors suggest that the inhibition of protein synthesis induce by them, is a result of the cleavage of the heteroduplex formed by PN and mRNA by an unknown enzyme [27].
The highly apical region of the 59-residues TAR stem-loop is a particularly good site for targeting by steric block oligonucleotides [28]. Synthetic molecules of PNA were used by Depecker and coworkers [29] to interact with the TAR RNA element of the HIV-1 genome. 2′-O-methyl, N3′-P5′-phosphorothioate and peptide nucleic acid targeted to TAR were shown to be efficient and sequence-specific inhibitors of HIV reverse transcription with IC50 in the nM range [30].
1.1.2.3 Delivery of Antisense Oligonucl...
Table of contents
- Cover
- Half Title page
- Title page
- Copyright page
- Preface
- List of Contributors
- Part 1: Nanomedicine
- Part 2: Drug Delivery and Therapeutics
- Index