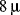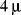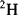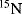![]()
Part I
Methodology
![]()
Chapter 1
Ionization Methods in Protein Mass Spectrometry
Ismael Cotte-Rodriguez, Yun Zhang, Zhixin Miao, and Hao Chen
Mass spectrometry (MS) has become one of the most powerful and popular modern physical-chemical methods to study the complexities of elemental and molecular processes in nature. The advent of new methods of ion generation, novel mass analyzers, and new tools for data processing has made it possible to analyze almost all chemical entities by MS, ranging from small organic compounds, large biological molecules, to whole living cells/tissues. As proteins fulfill a plethora of biochemical functions within every living organism, equally spectacular efforts and advances have been seen for protein ionization methods. In particular, the invention of matrix-assisted laser desorption ionization (MALDI) [1] and electrospray ionization (ESI) technologies [2, 3] allow one to measure protein molecular weights, to determine sequences, and to probe conformations and post-translational modifications of proteins. In addition the mass range of species amenable for MS analysis has been increased immensely, enabling the transfer into the gas phase of ionized noncovalent species with masses well over one million (e.g., a 100 MDa single DNA ion [4]). These advances move MS into the range of intact protein oligomers and functional machineries.
This chapter is an introduction to various ionization methods for proteins. As this is a broad topic with an immense literature coverage including many excellent books [5, 6] and reviews [7–17], we will emphasize some types of spray or laser-based protein ionization techniques, including atmospheric pressure MALDI, surface-enhanced laser desorption/ionization (SELDI), nanostructure-initiator MS (NIMS), sonic spray ionization (SSI), electrosonic spray ionization (ESSI), desorption electrospray ionization (DESI), fused-droplet electrospray ionization (FD-ESI), electrospray-assisted laser desorption ionization (ELDI), and matrix-assisted laser desorption electrospray ionization (MALDESI). We begin with the introduction of some historic facts for the development of protein ionization methods, followed with the description of each method including the ionization principles, strengths, and analytical applications.
1.1 History of the Development of Protein Mass Spectrometry
MS originates from ninteen-century physics. The first known mass spectrometer was built by J. J. Thomson in the early 1900s to study and measure the mass (m)-to-charge (z) (m/z) values of the “corpusules” that make up “positive rays” [18], a type of radiation initially observed by German physicist Eugen Goldstein. Following the seminal work of Thomson, MS underwent countless improvements in instrumentation, ionization methods, and applications. The classical ionization method, electron ionization (EI), was devised by Dempster and improved later by Bleakney [19] and Nier [20], and became a widely used standard for ionization of volatile organic compounds. This ionization technique requires extensive derivatization and evaporation of a nonvolatile analyte into the ion source, and it involves numerous fragmentation and rearrangement reactions.
Applications of MS to peptides (derivatized via acylation) begun in the late 1950s by Biemann [21] and McLafferty [22] The first methods that allowed analysis of nonderivatized peptides were field desorption (FD) and chemical ionization (CI) developed in the 1960s [23, 24]. Ionization by CI is achieved by interaction of its volatile molecules with reagent ions. CI allows ionization without significant degree of ion fragmentation but still requires gas-phase samples. Field desorption was reported by Beckey in 1969 [25], in which electron tunneling triggered by a very high electric field results in ionization of gaseous analyte molecules.
It was plasma desorption (PD) [26] and fast atom bombardment (FAB) [27] that opened the way to protein analysis. PD ionization, invented by R. D. Macfarlane in 1976 [28], a breakthrough in the analysis of solid samples, involves ionization of materials in the solid state by bombardment with ions or neutral atoms formed as a result of the nuclear fission of the Californium isotope
. In 1982 Sundqvist and coworkers obtained the first spectrum of a protein, insulin (
Figure 1.1), using bombardment with a beam of 90 MeV
ions from a tandem accelerator [26]. Later, FAB involving focusing the sample in liquid matrix with a beam of neutral atoms or molecules, was implemented for the ionization of proteins up to 24 kDa [29]. In 1983 Blakely and Vestal [30] introduced thermospray ionization (TSI) to produce ions from an aqueous solution sprayed directly into a mass spectrometer. Thermospray is a form of atmospheric pressure ionization in MS, transferring ions from the liquid phase to the gas phase for analysis. It was particularly useful in coupling liquid chromatography with mass spectrometry [31].
The breakthrough for large molecule laser desorption ionization came in 1987 when Tanaka combined 30-nm cobalt particles in glycerol with a 337-nm nitrogen laser for ionization and showed that singly charged protein molecular ions up to about 35 kDa can be introduced to a mass spectrometer [32]. During that time, MALDI [15, 33], first reported in 1985 by Hillenkamp, Karas, and their colleagues, emerged as the culmination of a long series of experiments using desorption ionization (DI). MALDI is a soft ionization technique for the analysis of biomolecules and large organic molecules and has gained wide success in protein analysis, particularly when coupled with time-of-flight (TOF) instruments [34, 35].
Another breakthrough occurred in 1984 when Fenn and coworkers used electrospray to ionize biomolecules [2]; the first ESI analyses of biopolymers including proteins were published in 1989 [3]. MALDI and ESI have revolutionized protein mass spectrometry since their invention in 1980s, and they have triggered the explosion in application of mass spectrometry for protein studies [36].
1.2 Laser-Based Ionization Methods for Proteins
1.2.1 Matrix-Assisted Laser Desorption/Ionization (MALDI)
Investigations of the wavelength influence in ultraviolet-laser desorption [33] led to invention of ultraviolet-laser matrix-assisted laser desorption ionization (UV-MALDI) between 1984 and 1986 and summarized in a 1987 paper [37]. In 1988 Karas and Hillenkamp reported ultraviolet-laser desorption (UVLD) of bioorganic compounds in the mass range above 10 kDa [1]. As a soft desorption ionization method, MALDI handles thermolabile, nonvolatile organic compounds, especially those with high molecular weight and can be successfully used for the analysis of proteins, peptides, glycoproteins, oligosaccharides, and oligonucleotides. Its operation is relatively straightforward, although matrix preparation requires experience and perhaps some artistry.
MALDI is based on the bombardment of sample molecules with laser light, process that allows sample ionization [38]. It requires a specific matrix consisting of small organic compounds (e.g., nicotinic acid) that exhibit a strong resonance absorption at the laser wavelength used. The sample is premixed and diluted with the highly absorbing matrix and allowed to dry on a sample target. A range of compounds is suitable as matrices: sinapinic acid is a common one for protein analysis while alpha-cyano-4-hydroxycinnamic acid is often used for peptide analysis (the structures of matrices are shown in Scheme 1.1). This kind of acid serves well as a matrix for MALDI owing to the acid's ability to absorb laser radiation and also to donate protons (H+) to the analyte of interest. Upon laser irradiation, energy is absorbed by the matrix in a localized region of the surface. As a result an explosive break up of the cocrystallized analyte/matrix sample occurs. The rapid expansion of the vaporized matrix in MALDI leads to the translational excitation of analyte molecules and the release of the analyte molecules from the surface of the condensed phase sample into vacuum. The analyte may be precharged (e.g., exist as a salt), and the intact analyte ion may simply be transferred as an ion from the solid to the vapor state upon laser irradiation of the matrix. Alternatively, a neutral analyte may be ionized through ion–molecule reactions (e.g., proton transfer reaction) occurring in the energized selvedge or interfacial region between the solid and gas phases.
MALDI has remarkable efficiency in producing intact molecular ions (often [M + H]+, [M + Na]+) of large biological compounds. MALDI ionization sensitivity is also extraordinary, and total amounts of sample loaded onto the target surface often are in the picomole to femtomole range. The method has tolerance to buffers and other additives and gi...