![]()
Section 1
Agrobacterium-Mediated Transformation
![]()
1
Host Factors Involved in Genetic Transformation of Plant Cells by Agrobacterium
Benoît Lacroix, Adi Zaltsman, and Vitaly Citovsky
Introduction
Agrobacterium tumefaciens and several other species of the Agrobacterium genus possess the unique ability to transfer a DNA segment from a specialized plasmid (tumor inducing or Ti plasmid in the case of A. tumefaciens and hairy root inducing or Ri plasmid for Agrobacterium rhizogenes, the two main species of pathogenic Agrobacterium) into a host plant cell. This feature is widely used in plant biotechnology, and Agrobacterium is, by far, the most important tool employed to produce transgenic plants (Newell 2000). Not surprisingly, the biology of Agrobacterium and its interactions with host plant have been the subject of numerous studies in the past three decades (for recent reviews, see Gelvin 2003; Citovsky et al. 2007; Dafny-Yelin et al. 2008).
In brief, the main steps of host genetic transformation mediated by A. tumefaciens are the following. The induction of Agrobacterium’s virulence machinery results in expression and activation of the virulence genes (vir genes) (Stachel et al. 1985b, 1986; McLean et al. 1994; Turk et al. 1994; Lee et al. 1996). This first step mobilizes a single-stranded DNA segment from the Ti or Ri plasmid. This segment of transferred DNA (T-DNA), delimited by two 25-bp direct repeat sequences known as left and right borders (LB and RB) (Peralta and Ream 1985; Wang et al. 1987), is termed the T-strand, and it represents the substrate of DNA transfer to the host cell. VirD2, associated with VirD1, forms a nuclease able to excise the T-strand by a strand-replacement mechanism, at the completion of which VirD2 remains covalently linked to the 5′-end (RB) of the T-strand (Ward and Barnes 1988; Young and Nester 1988; Durrenberger et al. 1989; Pansegrau et al. 1993; Jasper et al. 1994; Scheiffele et al. 1995; Relic et al. 1998). This VirD2–T-DNA complex is then translocated into the host cell cytoplasm by a mechanism relying on the VirB/VirD4 secretion system (Zupan et al. 1998; Vergunst et al. 2000; Christie 2004). The 11 proteins encoded by the VirB operon together with the VirD4 protein form a type IV secretion system, similar to the system allowing plasmid exchange by conjugation between bacteria. The type IV secretion system consists of a protein complex, spanning Agrobacterium internal membrane, periplasm and external membrane, and of an extracellular appendage, termed the T-pilus, composed mostly of VirB2 molecules forming a hollow channel (Christie et al. 2005). The VirB/VirD4 secretion system mediates the export of the VirD2–T-DNA complex out of the bacterial cytoplasm, and likely plays a role in its entry in the host cell. This secretion system is also required for the export of several Agrobacterium virulence proteins, that is, VirD5, VirE2, VirE3, and VirF, via their C-terminal secretion signals (Vergunst et al. 2000; Schrammeijer et al. 2003; Vergunst et al. 2003; 2005; Lacroix et al. 2005). There, the T-DNA–VirD2 complex is packaged by the single-stranded DNA-binding protein VirE2 (Christie et al. 1988; Citovsky et al. 1989; Sen et al. 1989). The resulting helical structure, called the T-complex, with the help of several bacterial and host proteins, is then imported into the host cell nucleus, targeted to the host chromatin, and ultimately integrated into the host genome (reviewed in Gelvin 2003; Lacroix et al. 2006a; Citovsky et al. 2007). The native T-DNA contains genes encoding enzymes that modify growth regulators and induce uncontrolled cell proliferation, which results in neoplastic cell growths (crown galls), and proteins mediating production and secretion of opines, amino acid, and sugar phosphate derivatives, secreted by the transformed cells and utilized almost exclusively by the Agrobacterium as carbon and nitrogen source (Escobar and Dandekar 2003).
The transfer of T-DNA is not sequence-specific, and any sequence of interest can be inserted between the T-DNA borders. The ability to engineer Agrobacterium to introduce genes of interest for plant genetic transformation is the basis of Agrobacterium’s use in biotechnology. The natural host range of Agrobacterium is very large, including most of the dicotyledonous and gymnosperm families (De Cleene and De Ley 1976). However, although the number of plant species transformable by Agrobacterium under laboratory conditions is always increasing (Newell 2000), in practice, producing transgenic plants efficiently is still a challenge for many plant species. Moreover, even nonplant species can be transformed by Agrobacterium under laboratory conditions (Lacroix et al. 2006b), including yeast (Bundock et al. 1995; Piers et al. 1996), various fungi (de Groot et al. 1998; Michielse et al. 2005), and cultured human cells (Kunik et al. 2001). This chapter focuses on numerous host plant factors that play important roles in the transformation process, from the initial interactions between Agrobacterium and plant cells and the activation of Agrobacterium’s virulence, to the integration of T-DNA into the host genome.
Plant Signals Affecting Agrobacterium’s Virulence Machinery
The rhizosphere is a complex and dynamic environment, where plant-associated bacteria such as Agrobacterium need subtle regulation systems to efficiently induce their virulence machinery (Brencic and Winans 2005). Agrobacterium’s virulence depends mostly on transcriptional activation of a set of virulence (vir) genes; this regulatory system allows the integration of environmental signals to ensure a timely expression of these genes. Moreover, the induction of virulence system obviously represents a high cost in energy for the bacterial cell, and its activation must be tightly regulated to ensure that it occurs only at the proximity of a susceptible host tissue. To this end, Agrobacterium harbors sensors able to recognize signals emitted by its host plants, and to activate the virulence machinery in response to these signals. The induction of vir gene expression in Agrobacterium relies on a two-component regulatory system encoded by the virA and virG genes that respond, directly or indirectly, to different plant and environmental cues (Klee et al. 1983; Stachel and Nester 1986). virA and virG have low basal expression, but their expression is highly inducible by a self-regulated system (Winans et al. 1988). The expression of other vir genes is virtually nonexistent in absence of induction, and it is strongly enhanced when the VirA–VirG system is activated. VirA–VirG represents a two-component regulatory system, in which VirA is the membrane-spanning sensor kinase that responds to external signals and activates the response regulator VirG by phosphorylation. Phosphorylated VirG recognizes and binds to a 12-bp long specific sequence, the vir box, which is present in all vir gene promoters, and serves to activate transcription (Brencic and Winans 2005).
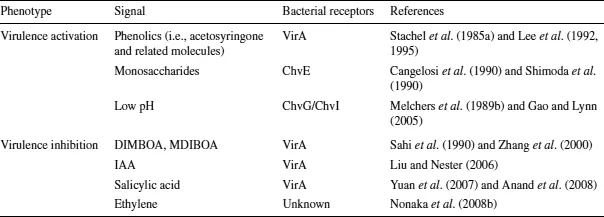
Several signals, from both host plants and the environment, can modulate vir gene expression (Table 1.1); these include phenolic compounds, monosaccharides, low pH, and low phosphate (McCullen and Binns 2006). Among these signals, only phenolics are absolutely required for virulence induction, whereas the other signals render Agrobacterium cells more sensitive to phenolics and/or enhance virulence induction levels.
Table 1.1 Plant and environmental signals that influence Agrobacterium virulence
Phenolic Compounds Activating Agrobacterium’s Virulence
Initially, during the analyses of plant cell exudates, a single phenolic compound, acetosyringone (3,5-dimethoxyacetophenone) was identified. It was present at elevated concentrations and able to induce vir gene expression even in the absence of the plant cells (Stachel et al. 1985a, 1986; Bolton et al. 1986). Since then, more than 80 related phenolics, including glycoside derivatives (Joubert et al. 2004), have been shown to act as vir inducers with variable efficiency (Melchers et al. 1989a; Palmer et al. 2004). These studies revealed that all vir-inducing molecules share common structural features that enable this family of chemicals to interact with bacterial receptors and to act as virulence inducers, suggesting that these molecules are recognized by a unique bacterial receptor (Lee et al. 1992). Whereas direct interaction between radioactively labeled acetosyringone and VirA has not been detected (Lee et al. 1992), genetic studies have demonstrated that phenolic inducers most likely interact directly with the linker domain of VirA, thereby activating VirA’s kinase activity (Lee et al. 1995). Indeed, the specific range of phenolic compounds recognized by different Agrobacterium strains was dependent on the virA locus, and could be transferred from one strain to another via the transfer of virA.
Reducing Monosaccharides
Sugar monomers are involved in vir gene activation in two ways: by enhancing VirA–VirG system sensitivity to phenols and by elevating the saturating concentration of phenols for virulence activation (Cangelosi et al. 1990; Shimoda et al. 1990). In addition, the range of phenolics recognized by the Agrobacterium vir gene induction system increases when monosaccharides are present as they act as coinducers (Peng et al. 1998). Several monosaccharides, such as D-glucose and D-galactose, are coinducers (Ankenbauer and Nester 1990; Shimoda et al. 1990), which share minimal structural features (i.e., the presence of a pyranose ring and acidic groups), also suggesting that they are recognized by a specific receptor. The virulence response to monosaccharides indeed relies on a chromosome-encoded factor, ChvE. This periplasmic sugar-binding protein is believed first to bind monosaccharides, then to interact with the periplasmic domain of VirA, and to enhance the VirA ability to activate vir gene expression (Cangelosi et al. 1990; Lee et al. 1992; Shimoda et al. 1993; Banta et al. 1994).
Low pH and Low Phosphate
Low pH (i.e., ∼5.7) enhances virulence activation, and this effect is mediated by VirA (Melchers et al. 1989b; Chang et al. 1996) as well as ChvE (Gao and Lynn 2005). Low pH and low concentration of phosphate (both are frequently observed in a variety of soils...