
eBook - ePub
In-cell NMR Spectroscopy
From Molecular Sciences to Cell Biology
- 296 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
In-cell NMR Spectroscopy
From Molecular Sciences to Cell Biology
About this book
In-cell NMR spectroscopy is a relatively new field. Despite its short history, recent in-cell NMR-related publications in major journals indicate that this method is receiving significant general attention. This book provides the first informative work specifically focused on in-cell NMR. It details the historical background of in-cell NMR, host cells for in-cell NMR studies, methods for in-cell biological techniques and NMR spectroscopy, applications, and future perspectives.
Researchers in biochemistry, biophysics, molecular biology, cell biology, structural biology as well as NMR analysts interested in biological applications will all find this book valuable reading.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access In-cell NMR Spectroscopy by Yutaka Ito, Volker Dötsch, Masahiro Shirakawa, Yutaka Ito,Volker Dötsch,Masahiro Shirakawa, Masatsune Kainosho in PDF and/or ePUB format, as well as other popular books in Physical Sciences & Biochemistry. We have over one million books available in our catalogue for you to explore.
Information
In-cell NMR Spectroscopy:
From 2001 to Now
CHAPTER 1
In-cell NMR Spectroscopy: From 2001 to Now
Institute of Biophysical Chemistry, Goethe University, Frankfurt, Germany
Email: [email protected]
1.1 In-cell NMR – The Basis
The dream of cell biologists is to observe their macromolecule of interest directly in its natural environment, inside a cell. Fluorescence microscopy has revolutionized the way proteins and nucleic acids can be visualized within this complicated, crowded surrounding. The advantage of fluorescence microscopy is its very high sensitivity and, in combination with super-resolution techniques, the ability to obtain high spatial resolution. This high sensitivity also allows for a high temporal resolution as the time it takes to detect signals with reasonable signal-to-noise ratios is short. However, detecting the localization of macromolecules in the cellular environment is not enough to understand their biological function. Interaction with other cellular components can only be obtained via fluorescence correlation microscopy, requiring two fluorescently tagged molecules. Interactions with a non-fluorescent molecule or conformational changes cannot be detected or only indirectly be proposed or require immunoprecipitation experiments. This gap in information can be – in principle – filled by NMR spectroscopy. During the last four decades, NMR spectroscopy has developed into a very important analytical tool in the chemical and biological sciences. In addition to its ability to determine structures of biological macromolecules under near-physiological conditions, NMR spectroscopy is capable of investigating the interaction between biological macromolecules and other molecules, ranging from proteins and nucleic acids to small organic compounds.
The ability to investigate intermolecular interactions is based on the sensitivity of the chemical shift to changes in the chemical environment. In addition to direct interaction with other molecules, changes in the chemical shifts of a biological macromolecule can also indicate changes to its conformation, for example different folding states. The development of in-cell NMR experiments that enable NMR investigations of biological macromolecules in their natural environment has created new tools for the direct investigation of the interaction between biological macromolecules and other macromolecules or drugs in living cells (Figure 1.1).1–7 In addition, these methods allow researchers to obtain information about the conformation and dynamics of macromolecules in their natural environment and to compare the in vivo behavior with data obtained from in vitro studies. A unique application of in-cell NMR is to study the folding and maturation of proteins directly following expression in the cellular environment.6,8,9
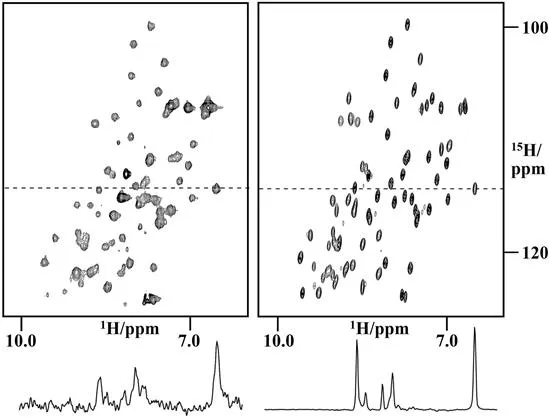
Figure 1.1 First in-cell NMR spectrum of NmerA in the cytoplasm of BL29 E. coli cells (left) and comparison with the spectrum of the purified protein (right).
1.2 Detecting the Molecule of Interest in the Crowded Environment of the Cell
The observation of biological macromolecules in cellular systems requires labeling schemes that enable the selective identification of these macromolecules in an environment that is crowded with other macromolecules as well as with many different small molecules.1,2,10–12 To make the macromolecule of interest visible in this crowded intracellular environment, macromolecules have to be labeled, either with NMR active isotopes or with paramagnetic probes for in-cell EPR studies.13–17 In principle, two different ways are possible to achieve labeling in the intracellular environment. The first method is the expression of the macromolecule of interest inside cells. The original in-cell NMR experiments used this overexpression method with bacterial cells,1,10,12 however, in the meantime overexpression has also been achieved with other cell types including different eukaryotic cells (yeast,18 insect cells,19 mammalian cells6). Overexpression, particularly in bacteria, is convenient and uses very well established protocols, but it has the potential of producing a huge amount of background signals since not only the macromolecule of interest but all cellular components will be labeled with NMR-active isotopes. The original in-cell NMR experiments in bacteria surprisingly demonstrated that uniform 15N-labeling of the overexpressed protein is feasible and that only a minimal level of background signals is produced.1,10 The only requirements for observing an overexpressed 15N-labeled protein in in-cell NMR experiments are that its expression level reaches a certain threshold and that its rotational correlation time is sufficiently short.10 The last condition is met in principle for all macromolecules that can also be investigated by NMR in vitro and that do not interact with large cellular components since the intracellular viscosity is only a factor of two higher than the viscosity of water.20,21
In contrast, uniform labeling of overexpressed macromolecules with 13C results in a high level of background signals that makes unambiguous identification of the overexpressed macromolecule impossible.22 One exception is signals with unique chemical shifts such as high field shifted methyl groups or anomeric sugar resonances.12,22 The difference in the level of background signals between 15N and 13C was attributed to the fact that C–H groups are far more abundant in small molecules in the cell than N–H groups, and that many amide protons in small molecules are not protected from solvent exchange, thus making their NMR signals undetectable due to exchange line broadening.10
To overcome this problem of high background signals, amino acid-type selective labeling schemes have been suggested and successfully applied.22 In particular in bacterial cells, the use of isotopically labeled amino acids for selective labeling schemes has to be considered carefully, as amino acids often represent precursors for the synthesis of other amino acid types, thus leading to isotopic scrambling. Amino acids that are at the end of a metabolic pathway have been shown to be useful for in-cell NMR experiments such as Lys, Arg or His for 15N labeling and Met and with some restriction Ala for 13C labeling. Another strategy is to use labeled precursors, for example α-ketobutyrate to incorporate labeled isoleucine into the protein of interest in bacteria.23 Likewise, the use of fluorinated amino acids for labeling of overexpressed proteins has been successfully used and produced background-free spectra.24,25
A different strategy that was proposed is to suppress the high background by selective expression of the protein of interest with the concomitant suppression of the expression of all other (host) proteins. This suppression of host gene expression can be achieved by two different techniques. The first is based on using the drug rifampicin, which inhibits the bacterial RNA polymerase.26 The RNA polymerase of the bacteriophage T7, however, is not affected by this drug, thus allowing the selective overexpression of the protein of interest from a plasmid containing a T7 promoter in bacterial cells expressing the T7 RNA polymerase. The second approach is degradation of all mRNA molecules that contain ACA sequences by an mRNA interferase.27 Changing all ACA base triplets in the DNA sequence of the protein of interest by utilizing other codons enables the selective translation of the mRNA of a particular protein while all other mRNAs that contain ACA triplets are degraded.
1.3 In-cell NMR with Eukaryotic Cellular Systems
While these initial bacteria-based in-cell NMR experiments were used to study different aspects of protein behavior, for example protein–protein interactions as well as protein interactions with small molecules, it was clear from the beginning that the method had to be applied to eukaryotic cell systems as well. For these systems, new methods to get isotopically labeled macromolecules into the cellular interior were developed. The first one used is based on micro-injecting a highly concentrated sample of a purified macromolecule labeled with NMR-active isotopes into the cell type of interest.28,29 This method, however, is res...
Table of contents
- Cover
- Title
- Copyright
- Preface
- Contents
- Part 1: In-cell NMR Spectroscopy: From 2001 to Now
- Part 2: Host Cells for In-cell NMR Studies
- Part 3: Basic In-cell NMR Techniques
- Part 4: In-cell NMR Applications
- Subject Index