![]()
CHAPTER 1
Design of In Situ Cytosensing Strategies
1.1 General Principles of In Situ Cytosensing
Owing to the continuing attention devoted to human health and life, recent decades have witnessed tremendous developments in analytical strategies for the in situ probing of chemical and biological events at the molecular level in living cells. These tools provide spatiotemporal information on cellular molecules, biochemical reactions and signalling and communication processes,1–3 thus contributing to the better understanding of the sophisticated mechanisms of life and to improvements in or even transformation of disease diagnosis and treatment concepts and techniques.4,5 To analyse cellular functional molecules in situ, one needs to design a detection system composed of a recognition module, a signal transduction module and a signal output module. The recognition module achieves the differentiation of the targets by specific binding and the signal transduction module converts the recognition event into a signal, transformation or reaction to indicate the existence of the target. The latter can be integrated with the capability to amplify the signal or implement multiplexed analysis. The last module performs the final collection and reporting of the signals that correspond to the target. Owing to the low expression levels of the targets and the complex biological environment, four key aspects should be taken into consideration when designing the system: recognition/binding specificity, signal generation pathway, signal amplification and multi-channel analysis capability. These issues play crucial roles in the analytical performance and the comprehensive consideration of these aspects is challenging. This chapter presents a review of the most promising trends regarding these major concerns.
1.2 Enhancement of Detection Specificity
Specific detection depends on the recognition of a biosensing interface towards the target. Common recognition motifs include antibodies, lectins, peptides, nucleic acids (including aptamers), biomimetic polymers, artificial receptors and small molecules. To improve the detection sensitivity, it is beneficial to use recognition motifs with high specificity and avidity towards the target, which are often intrinsic properties of the individual recognition pairs. In spite of this, one can still tailor the detection schemes to enhance the binding intensity and specificity. For example, to improve the avidity of lectins towards their corresponding glycan epitopes, the glycans can be assembled in a multivalent fashion to accelerate the binding events. Based on this strategy, Ding et al. fabricated a mannan-modified gold electrode to yield a carbohydrate monolayer with multivalent binding capability with the lectin concanavalin A (Con A), thus permitting efficient competition between the mannan monolayer and cell-surface mannose.6
To increase the specificity for the identification of a given type of cells, Rudchenko et al. developed automata to perform cascade strand-displacement reactions directed by antibody recognition towards cell-surface markers (Figure 1.1).7 The automata checked the existence of each marker successively according to an “if yes then proceed” logic. Only when all the markers of the given set were present on the cell surface could the automata complete the computation and output a final fluorescence signal, thus achieving the identification of a specific population of lymphocytes within human blood cells.
Figure 1.1 Scheme of automata operating on a B cell with a C45+CD20+ phenotype (target) and on an example of a non-targeted cell with a CD45+CD20− phenotype.7 Reproduced from ref. 7 with permission from Springer Nature, Copyright 2013.
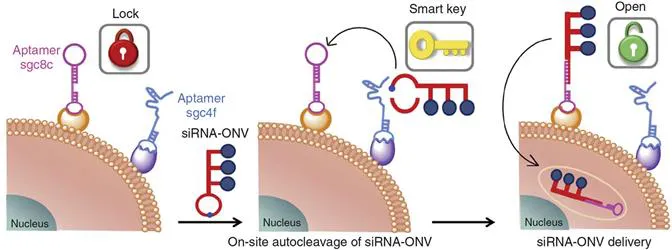
Concerning the targeting delivery, most current cell targeting strategies rely on only one receptor on the cell surface and therefore usually suffer from the issue of high non-specific interactions. The incorporation of two factors in a targeting system to function in a serial manner may improve the site-specific delivery of the probe. On the basis of this concept, Ren et al. used an auto-cleavable hairpin structure acting as “smart keys” to modify the siRNA-loaded oligonucleotide nanovehicle (siRNA-ONV) and bound two kinds of aptamers, sgc8c and sgc4f, on the cell surface to act as “double locks” (Figure 1.2).8 These “locks” could be opened sequentially by reacting with the “key” in a serial manner. The “dual lock-and-key” mode controlled the cell “locked-open” status and thus achieved cell-subtype-specific recognition and precise probe delivery.
Figure 1.2 Scheme of the working principle of the siRNA-ONV nanotube.8 Reproduced from ref. 8, https://doi.org/10.1038/ncomms13580, under the terms of the CC BY 4.0 licence, http://creativecommons.org/licenses/by/4.0/.
In addition to dual specificity control of the same type, different types of specificity control can also be integrated into one system. For intracellular sensing, considering the complex intracellular environment, it is beneficial to turn off the response capability of the recognition motif of the detection probe until it reaches the desired locations, otherwise the recognition and the subsequent signal generation may take place during the cellular delivery and uptake process, leading to impaired detection accuracy and low signal-to-noise ratio. In this context, Zhao et al. designed a nanodevice containing two dependent components: a UV light-activatable DNA aptamer probe and lanthanide-doped upconversion nanoparticles (UCNPs).9 The Cy3-modified aptamer strand was initially locked by a quencher-bearing complementary DNA containing a photocleavable (PC) group, which was denoted a “PC inhibitor”, hence the aptamer could not bind to ATP in its locked state and displayed a low fluorescence signal background. When the nanodevice reached the desired region, upon irradiation with near-infrared (NIR) light the PC inhibitor would be cleaved at the PC site by the UV light converted by UCNPs and the capability of the aptamer to switch its structure to bind ATP would be restored. Thus, in the presence of ATP, the dissociation of the cleaved PC inhibitor led to a substantial increase in the fluorescence signal, achieving ATP sensing with high specificity.
1.3 Design of Off–On Signal Switch
When the recognition motif for the target is fixed, a signal transduction pathway must be designed to indicate the occurrence of the recognition/binding events. To achieve high-sensitivity detection and to lower the background signals, a key point is the design of a signal off–on switch that can be specifically triggered by the target or target-associated events to reflect the existence of the target in the analysis scheme. The tailoring of signal generation methods depends on the action modes of the recognition processes, for example, cleavage by an enzyme or hybridization with a DNA strand.
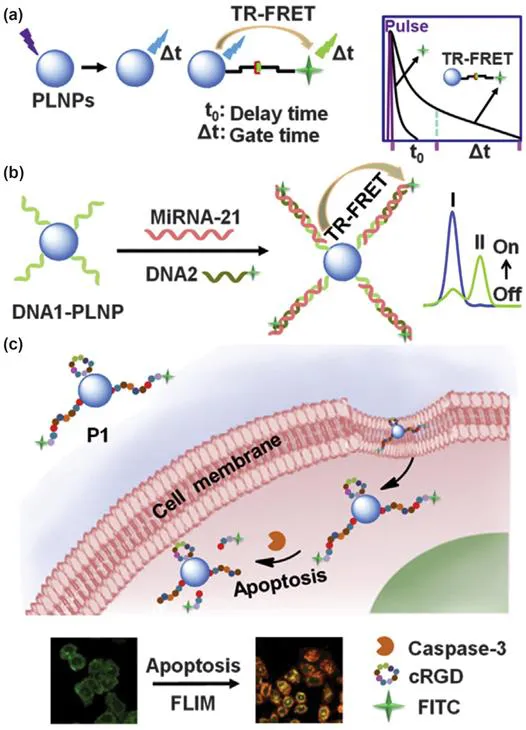
With regard to cellular enzymes with cleaving capability, it is convenient to design signal switches by use of responsive substrates. One can label the substrate with a fluorophore at one end and use the other end to link to a nanomaterial with desired optical properties, thus constituting a certain type of energy transfer pathway, such as fluorescence resonance energy transfer (FRET). The reason for the introduction of nanomaterials lies in the synergistic employment of their unique characteristics not only from an optical properties perspective but also with regard to the accelerating capability for cellular delivery.10,11 However, FRET-based detection platforms often suffer from a high background signal due to acceptor bleed-through. A smart solution for this issue is the design of a time-resolved fluorescence resonance energy transfer (TR-FRET) system using persistent luminescence nanoparticles (PLNPs), in which the luminescence of the fluorophore is collected only after a delay time, thus suppressing the acceptor bleed-through. Zhang et al. demonstrated this concept by developing a TR-FRET platform for the in situ lifetime quantification of intracellular caspase-3 (Figure 1.3).12
Figure 1.3 Schematic illustration of (a) the PLNP-based TR-FRET principle, (b) TR-FRET-based detection of miRNA-21 using caspase-specific DNA1-functionalized PLNPs and (c) lifetime imaging of intracellular caspase-3 activity during cell apoptosis. The blue and green lines in (b) represent the time-resolved fluorescence curves before and after response to the target, respectively.12 Reproduced from ref. 12 with permission from Elsevier, Copyright 2015.
Another common design for enzymes with cleaving capability is to load a peptide-based fluorophore-labelled substrate on nanomaterials with fluorescence quenching ability, such as graphene oxide (GO),13 gold nanorods (AuNRs),14,15 manganese dioxide16 or a metal–organic framework (MOF).17 The cleavage by the corresponding enzymes leads to a signal-on switch by releasing the fluorophore into solution. For example, Zhang et al. fabricated a caspase-3-responsive multifunctional nanoprobe by assembling a porphyrin, a folate targeting motif and a dye-labelled peptide substrate in an MOF cage. Upon executing enhanced photodynamic therapy (PDT), owing to the integration of porphyrin and MOF, this probe allows in situ cell apoptosis monitoring by displaying caspase-3 activation-induced fluorescence recovery, thus underlining its great promise in precision cancer therapy.17
Cathepsin B (CaB) is another analyte that has attracted much attention owing to its crucial roles in cancer progression and diagnosis, and displays a specific peptide-cleaving capability.18 Taking CaB as the target, Tian et al. developed a multi-functional theranostic nanoplatform by the non-covalent assembly of 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-{folate[poly(ethylene glycol)]-2000} (DSPE-PEG2000-FA) and chlorin e6 (Ce6)-labelled CaB-specific peptide substrate (Ce6-G...