![]()
CHAPTER 1
Lipidomics Basics
W. J. Griffiths*a , E. Yutuc a , D. Davies a , A. Dickson a , R. Angelini a , D. El Assad b , G. Frache b and Y. Wang a
a Swansea University Medical School, ILS1 Building, Singleton Park, Swansea, SA2 8PP, UK,
b Luxembourg Institute of Science and Technology, 41, rue du Brill, L-4422 Belvaux, Luxembourg
*E-mail:
[email protected] Lipidomics can be regarded as the quantitative profiling of the entire lipid composition of a defined system, be that a cell, tissue, biofluid or intact organism. Lipidomics is a descendent of what was previously known as “metabolite profiling” and is a relative of the related “omic” disciplines of genomics, transcriptomics and proteomics. Lipidomics can be regarded as a sub-division of metabolomics. In this chapter we will discuss the current methodologies popular in lipidomics, highlighting gas-chromatography mass spectrometry (GC-MS), liquid-chromatography MS (LC-MS) and direct-infusion, also known as “shotgun” MS (DI-MS). We will also introduce newer methods including low-flow-rate-LC MS, ion mobility MS (IM-MS) and MS imaging (MSI), described in detail in later chapters. Chemical derivatisation is not a new idea to lipidomics; however, its use goes in and out of vogue, hence we also introduce some of its benefits and disadvantages here and in a later chapter.
1.1 Introduction
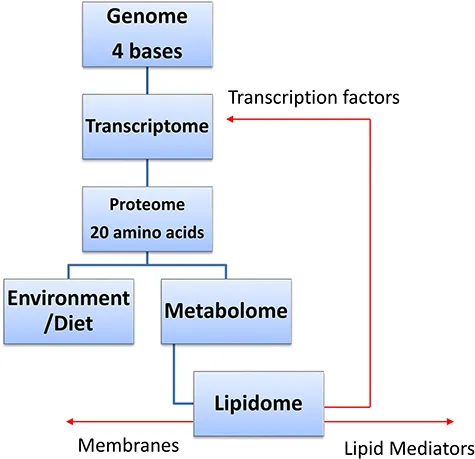
The term lipidomics was introduced at around the turn of the century. 1,2 It refers to the quantitative profiling of the lipidome, which is the lipid composition of a system. The lipidome is a subdivision of the metabolome which includes all the small molecules, usually thought of as having a mass < 1500, in a system (Figure 1.1). Both lipidomics and metabolomics are modern versions of “metabolite profiling” introduced as far back as the early 1970s. 3 Currently, the methods of lipidomics are dominated by the use of mass spectrometry (MS) although nuclear magnetic resonance spectrometry (NMR) can also be used. 4 The most popular ionisation method for MS-based lipidomics is electrospray-ionisation (ESI) which is readily linked with liquid-chromatography (LC) separations and with direct-infusion (DI) or “shotgun” sample-introduction formats. Other ionisation methods include desorption-ESI (DESI) 5 and matrix-assisted laser/desorption ionisation (MALDI), 6 both widely used for MS imaging (MSI) and electron ionisation (EI) incorporated in most gas-chromatography (GC)-MS methods. 7
Figure 1.1 The lipidome in relation to other biological “omes”.
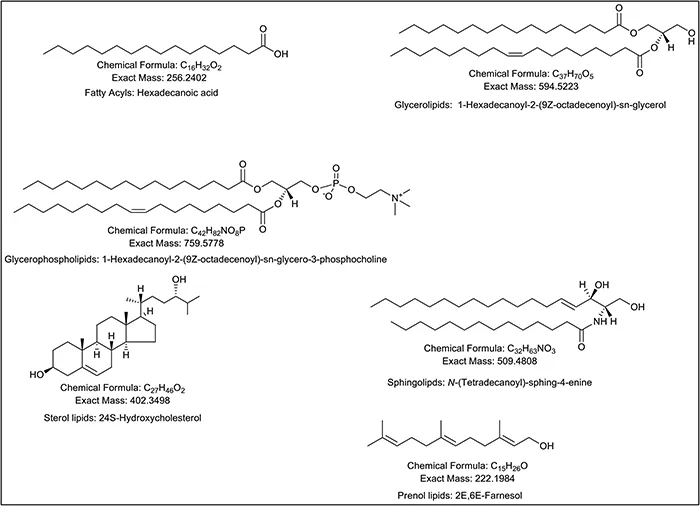
The lipidome is extremely complex with eight different categories, six of which dominate the mammalian lipidome (Figure 1.2) 8 and lipidomics' methods are highly dependent on the nature of the problem to be solved. If hundreds, or even thousands of samples are to be analysed in a study, then it is most practical to perform only a minimum of sample pre-treatment and to perform rapid MS analysis using DI-MS (e.g. ref. 9 and 10). On the other hand, if the object of the study is to determine the most comprehensive measurement of a lipidome, with only a few samples to be analysed, then extensive sample preparation protocols followed by multiple analysis may be the best route. 11,12 The reader is directed to the Lipid Maps website, specifically http://www.lipidmaps.org/resources/protocols/, for protocols for the analysis of different lipid categories.
Figure 1.2 Six main categories of lipids.
1.2 Most Common Experimental Methods in Lipidomics
1.2.1 DI and Shotgun Methods
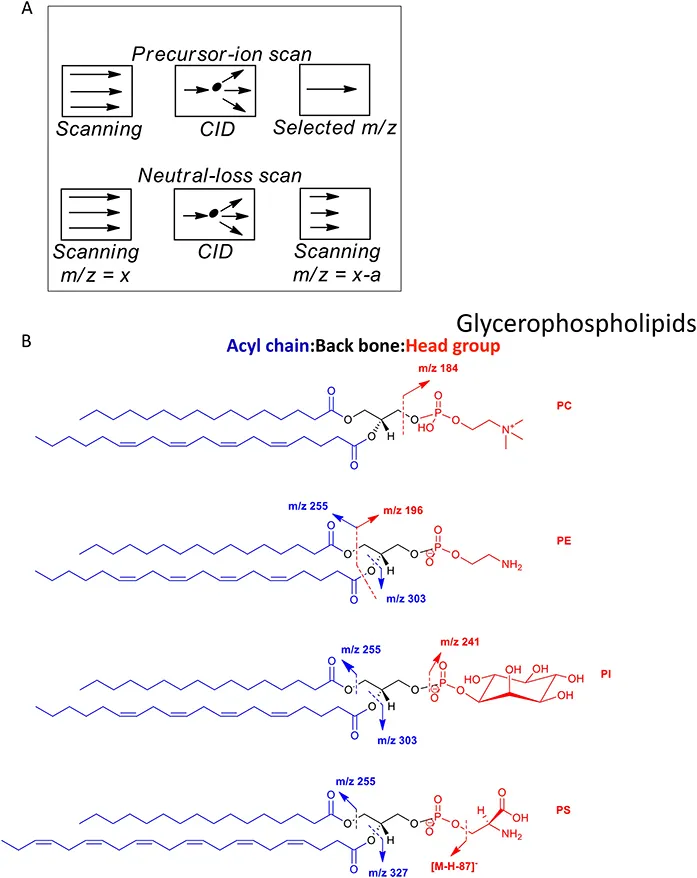
The terms “shotgun” and abbreviation DI are essentially interchangeable according to current lipidomic terminology. Simply, a sample is infused into the ESI source of a MS in the absence of chromatography. This may be achieved by injecting a sample from an LC-autosampler, in the absence of a column, via a syringe pump, via a nano-ESI needle or via a chip-based nano-ESI nozzle as commercialised by Advion Inc. The method then relies on the MS to perform the identification. This, as initially exploited by Han and co-workers, may be realised by utilising multiple consecutive scan modes on a tandem quadrupole MS (Figure 1.3) 1,2,13 or using high-resolution scans often on Orbitrap instruments 9,14 using both positive- and negative-ion modes. Lipids fragment in tandem mass spectrometry (MS/MS) and multistage fragmentation (MS n ) experiments according to well established ion-chemistry rules, allowing the prediction of fragment ion spectra, a significant advantage for compound identification in DI-MS(MS/MS) methods. 15,17 The interested reader is directed to the text “Tandem Mass Spectrometry of Lipids: Molecular Analysis of Complex Lipids” by Dr Robert Murphy, one of the pioneers of modern lipid analysis. 18 The major advantages of DI-MS methods are simplicity in sample handling and speed of sample analysis. The disadvantage is that DI-MS spectra are dominated by the most abundant or easily ionised lipids. This limitation can be overcome by performing dedicated sample preparation protocols targeted at one class of analytes at a time 19 or by performing specific derivatisations targeting a defined functional group. 20
Figure 1.3 (A) Important MS/MS scan modes utilised in DI-MS/MS studies performed on tandem quadrupole instruments. See Chapter 10. (B) Lipids fragment according to well established rules of ion chemistry. Abbreviations: PC, phosphocholine (glycerolphosphocholine); PE, phosphoethanolamine (glycerophosphoethanolamine); PI, phosphoinositol (glycerophosphoinostol) and PS, phosphoserine (glycerophosphoserine).
1.2.2 LC-MS Methods
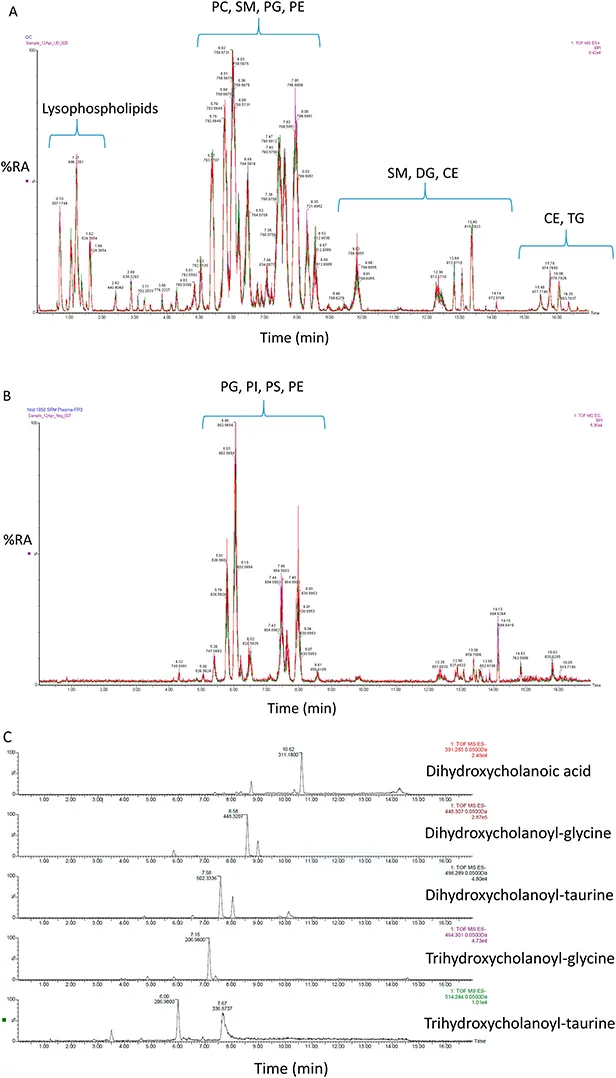
The advantage of utilising LC is that it can provide molecular separation, the penalty of its use is that it consumes time. LC-MS for lipidomics is often performed using reversed phase (RP) or hydrophilic interaction liquid chromatography (HILIC) columns that separate according to chain length and head group polarity, respectively. 21,22 The two stationary phases can be utilised in a two-dimensional-LC (2D-LC) format, with the first separation according to the hydrophobic portion of the molecules (RP column) followed by separation of lipid classes based on polarity (HILIC column). 23 LC separations with RP and HILIC columns may take up to 1 hour, although application of ultra-performance-LC (UPLC), with chromatography performed at higher pressures, can reduce the run time (Figure 1.4). Both RP and HILIC utilise solvent systems compatible with ESI. This is not generally so with normal phase (NP) chromatography where alternative ionisations methods, e.g. atmospheric pressure chemical ionisation (APCI) may be more appropriate.
Figure 1.4 (A) UPLC-MS separation of lipids in the NIST SRM1950 of human plasma in (A) positive-ion mode and (B) negative-ion mode. (C) Bile acid analysis in the negative-ion mode after solid phase extraction. Data generated on an Acquity UPLC, with a 2.1 mm × 100 mm C18 column exploiting a Xevo-G2-XS quadrupole time-of-flight (QTOF) MS (Waters Corporation). In (A) and (B) the reproducibility of chromatography is demonstrated by three overlaid chromatograms. Abbreviations: PC, phosphocholine (glycerolphosphocholine); SM, sphingomyelin (phosphosphingolipid); PG, phosphoglycerol (glycerophosphoglycerol); PE, phosphoethanolamine (glycerophosphoethanolamine); DG, diacylglycerol (diradylglycerol); CE, cholesterol ester (sterol ester); TG, triacylglycerol (triradylglycerol); PI, phosphoinositol (glycerophosphoinostol) and PS, phosphoserine (glycerophosphoserine). The NIST SRM1950 is used as a standard reference material (SRM) in lipidomics. 12,24,25
1.2.3 GC-MS Methods
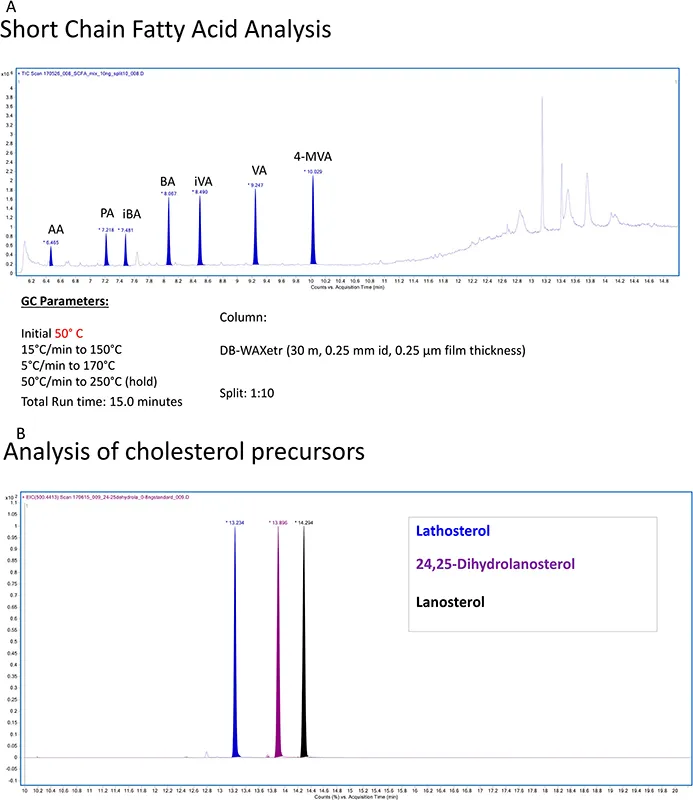
GC-MS has been used for the profiling of lipids for decades. 26,28 It is still the ideal method for measuring underivatised short chain fatty acids and is also widely used for analysing fatty acids as their methyl esters (Figure 1.5). 29,30 With the exception of small volatile compounds, a disadvantage of using GC-MS in lipidomics is the requirement for derivatisation to enhance volatility and stability. There is a huge volume of literature describing derivatisation reactions appropriate for lipids. The interested reader is directed to the classical books by Blau and King, 31 Blau and Halket 32 and more recent reviews by Halket and Zaikin. 33 GC has the advantage over LC of providing a greater number of theoretical plates, hence superior resolution. GC-MS is extensively exploited for steroid and sterol analysis where robust and well documented analytical protocols are in place (Figure 1.5B). 34,36
Figure 1.5 (A) Analysis of short-chain fatty acids in the absence of derivatisation by GC-MS. The column was a DB-WAXetr, the run time was 15 min and the temperature gradient was as follows: initial temperature 50 °C, raised to 150 °C at 15 °C mi...