
Medical Biotechnology
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Medical Biotechnology
About this book
The future is now—this groundbreaking textbook illustrates how biotechnology has radically changed the way we think about health care
Biotechnology is delivering not only new products to diagnose, prevent, and treat human disease but entirely new approaches to a wide range of difficult biomedical challenges. Because of advances in biotechnology, hundreds of new therapeutic agents, diagnostic tests, and vaccines have been developed and are available in the marketplace.
In this jargon-free, easy-to-read textbook, the authors demystify the discipline of medical biotechnology and present a roadmap that provides a fundamental understanding of the wide-ranging approaches pursued by scientists to diagnose, prevent, and treat medical conditions.
Medical Biotechnology is written to educate premed and medical students, dental students, pharmacists, optometrists, nurses, nutritionists, genetic counselors, hospital administrators, and individuals who are stakeholders in the understanding and advancement of biotechnology and its impact on the practice of modern medicine.
Hardcover, 700 pages, full-color illustrations throughout, glossary, index.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Information
SECTION I
The Biology behind the Technology
- 1 Fundamental Technologies
- 2 Fundamental Concepts in Immunology
- 3 The Genetic Basis of Disease
- 4 Immune Pathogenesis
- 5 Microbial Pathogenesis
1
Fundamental Technologies
- Molecular Cloning
- Preparation of DNA for Cloning
- Insertion of Target DNA into a Plasmid Vector
- Transformation and Selection of Cloned DNA in a Bacterial Host
- Cloning Eukaryotic Genes
- Recombinational Cloning
- Genomic Libraries
- Amplification of DNA Using PCR
- DNA Sequencing Technologies
- Dideoxynucleotide Procedure
- Pyrosequencing
- Sequencing Using Reversible Chain Terminators
- Sequencing by Ligation
- Sequencing Whole Genomes
- Shotgun Cloning Strategy
- High-Throughput Next-Generation Sequencing Strategies
- Genomics
- Transcriptomics
- Proteomics
- Metabolomics
- SUMMARY
- REVIEW QUESTIONS
- REFERENCES
Molecular Cloning
Preparation of DNA for Cloning
DNA
mRNA
box 1.1 The Development of Recombinant DNA Technology
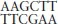
Table of contents
- Cover
- Table of Contents
- Preface
- About the Authors
- SECTION I: The Biology behind the Technology
- SECTION II: Production of Therapeutic Agents
- SECTION III: Diagnosing and Treating Human Disease
- Glossary
- Index
- End User License Agreement