
- 336 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Molecular and Cellular Methods in Developmental Toxicology
About this book
This new manual provides a convenient source of experimental procedures, including the most modern and frequently used molecular and cellular techniques. Experimental protocols have been carefully selected by developmental toxicologists for developmental toxicologists. The most important new trends, such as evaluation of the safety of therapeutic antisense oligonucleotides, studies of the role of cell death in abnormal development, and the identification of sparingly expressed developmental control genes are featured. This is the perfect manual for scientists trained in classical developmental toxicology who want to add molecular and cellular methods to their research.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Molecular and Cellular Methods in Developmental Toxicology by George P. Daston in PDF and/or ePUB format, as well as other popular books in Medicina & Teoria, pratica e riferimenti medici. We have over one million books available in our catalogue for you to explore.
Information
Part I
Molecular Techniques
Chapter Molecular Techniques for Developmental Toxicology: An Introduction | 1 |
Contents
I. Molecular Biology: The Basics
II. Some General Rules
III. Preparing Probes
A. Restriction Enzyme Cut of DNA
B. Isolation of cDNA Fragment
C. Elution from Low Melting Point Agarose
D. Filling in Cut Ends
E. Ligating the Insert and Vector
F. Making Probes
IV. RNA Extraction
V. Agarose Gel Electrophoresis
VI. Detecting Specific mRNAs
VII. Northern Blotting
A. Transfer to Nylon Membrane
B. Hybridization of RNA
C. Posthybridization
VIII. Slot Blotting
IX. RNase Protection Assay
X. Use of Oligonucleotides
XI. Other Resources
Appendix A: Reagents Used in Molecular Biology
Appendix B: Partial List of Suppliers of Molecular Biology Products and Equipment
The technology of molecular genetics has revolutionized research in a large number of fields, including developmental toxicology. It is now possible to directly address questions about the effect of xenobiotic chemicals on the expression and function of the genes controlling development. This ability to conduct research at the most basic level of biological organization is already dramatically changing our understanding of the control mechanisms of development and how they are perturbed, leading to congenital malformations.
The armament of molecular techniques is truly vast and beyond the scope of this chapter or even this book. There are comprehensive methods anthologies available, and these are listed at the end of this chapter. In recent years, kits have been assembled that provide the investigator with a complete set of reagents and written procedures for accomplishing a specific task. I have not provided extensive protocols for procedures that can be better done by using a kit. A list of suppliers of molecular biology equipment, supplies, and kits is given in Appendix B.
I. Molecular Biology: The Basics
Deoxyribonucleic acid (DNA) is the genetic material of cells. It encodes all of the information specifying the primary structure of all proteins in the organism. DNA is a double-stranded polymer. Each strand consists of four nucleotides — adenine (A), guanine (G), cytosine (C), and thymine (T) — linked by a backbone of covalently linked deoxyribose sugars and phosphoesters. The two strands are held together by hydrogen bonding between the nucleotides of opposite strands. This bonding is specific and complementary: A binds T, C binds G. The strands are twisted around each other (the famous double helix) and are in opposite polarity with respect to their phosphate-sugar backbones.
DNA serves as a template for making ribonucleic acid (RNA), which in turn serves as the template for the synthesis of proteins. RNA is similar to DNA except that it is single stranded, has ribose instead of deoxyribose, and has uridine (U) instead of thymine. We will be most interested in evaluating mRNA (m for messenger), because its expression dictates the kinds of proteins that a cell synthesizes and is therefore cell type and developmental stage specific. However, there are other types of RNA, particularly ribosomal (r) and transfer (t) RNA, that compose the majority of the RNA in a cell. Even though the RNA is single stranded, it will complex with a complementary strand of DNA or RNA. This makes it possible to use sequences of DNA or RNA to detect specific mRNA.
The genes in prokaryotic organisms are arranged in a continuous linear fashion such that the DNA sequence corresponds directly to the mRNA sequence. This simple arrangement is the key to many molecular techniques. Eukaryotic genes on the other hand are interspersed with regions called introns that are not translated into proteins. The regions that contain the coding sequence for proteins are called exons. In eukaryotes, a RNA transcript of both exons and introns is first synthesized, and then the introns are removed and the exons joined together to form mRNA. In a final processing step, almost all mRNA has a poly-A tail added to the 3′ end before it leaves the nucleus. This unique feature can be utilized to separate mRNA from total RNA when that is desirable, by techniques that will be described later in the chapter.
One of the most important early discoveries of molecular biology was that, in addition to DNA serving as a template for RNA, certain viruses were able to make DNA from a RNA template using the enzyme reverse transcriptase. The DNA made in this way is called cDNA (c for complementary). Bacterial restriction enzymes are another important discovery in molecular genetics. These enzymes cut DNA at specific nucleotide sequences. Hundreds of these enzymes have been discovered, and a large number of target sites are available. It is possible to pick and choose among these to cut out a specific region of DNA whose sequence is known. It is also possible to cut the entire genome into manageable pieces for the construction of a library of DNA.
Libraries of DNA can be constructed from the entire genome of an organism by treating cellular DNA with restriction enzymes (genomic library). Or, a library of cDNA made by reverse transcriptase can be made only from the mRNA being expressed by a particular cell type or tissue. In either case, the DNA is inserted into vectors, which are DNA molecules that can be introduced into a host cell (usually a bacterium) where the vector along with its insert can be propagated. This procedure is known as cloning, and the host cell is called a clone. The most commonly used vectors are plasmids, circular pieces of DNA that are replicated extrachromosomally in bacteria. Plasmids have been engineered so that they have multiple features that make it easy to insert and transcribe cDNA and to select clones that contain a plasmid. A typical plasmid vector, marketed by Stratagene, is shown in Figure 1.1. It contains:
1. A multiple cloning site, with a large number of different restriction enzyme recognition sites to facilitate the introduction or removal of the cDNA
2. An antibiotic resistance gene, so that only the bacteria that have been transformed by introduction of a plasmid can survive antibiotic treatment
3. A lac Z (β-galactosidase) gene, to allow for blue-white selection: the gene is interrupted by introduction of DNA into the multiple cloning site, and the β-galactosidase protein produced is nonfunctional; the clones can be incubated in the presence of a colorless substrate that is made into a blue product by functional β-galactosidase, allowing one to determine whether the plasmid has a DNA insert; many vectors including this one also contain a lac I repressor gene that can be chemically inhibited to enhance the contrast between blue and white colonies
4. Promoter regions flanking the insert site: this feature allows RNA polymerase to transcribe either strand of the cDNA (the two promoters bind different polymerases) and allows the synthesis of RNA that is either complementary (anti-sense) or identical (sense) to the original mRNA from which the clone was derived
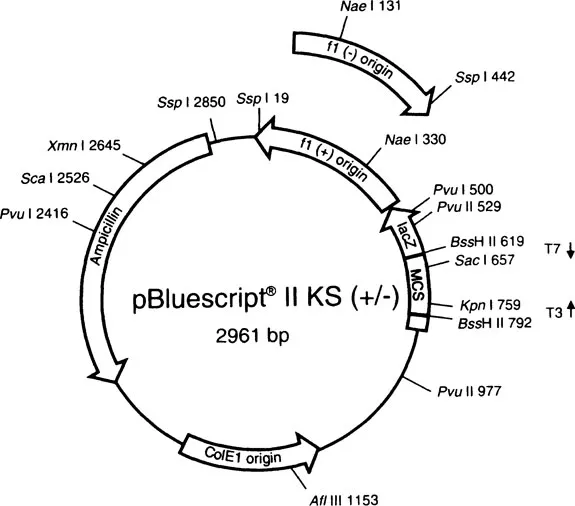
FIGURE 1.1
A typical plasmid vector. It has a multiple cloning site (MCS), an ampicillin resistance gene, a lac Z gene for blue-white selection, and promoters flanking the insertion site (T3 and T7 for T3 and T7 RNA polymerases) so that both strands of cDNA can be transcribed. This plasmid is sold by Stratagene. (Figure courtesy of Stratagene Technical Services.)
A typical plasmid vector. It has a multiple cloning site (MCS), an ampicillin resistance gene, a lac Z gene for blue-white selection, and promoters flanking the insertion site (T3 and T7 for T3 and T7 RNA polymerases) so that both strands of cDNA can be transcribed. This plasmid is sold by Stratagene. (Figure courtesy of Stratagene Technical Services.)
In addition to these features, plasmids are easy to isolate from the host bacteria and are easy to sequence. Disadvantages are that it is still tedious to screen plasmid libraries and the length of the insert must be relatively short, less than 10 kilobases (Kb) of DNA. Other vectors that are often used include bacteriophage lambda, cosmids (hybrids of phage and plasmid), and yeast artificial chromosomes, which can accept inserts of 1000 Kb or more. For the purposes of generating large quantities of RNA probes from cDNA that has already been isolated, however, plasmids are ideal.
Most novices will not be involved in cloning and sequencing libraries, so protocols for these will not be included here. Perhaps the most widely applicable techniques for developmental toxicology are those for measuring the expression of specific mRNAs. These techniques use cRNA probes that are transcribed from plasmids. The procedures for making the probes vary somewhat depending on the identity of the promoter region, so there is no generic protocol provided. Most vendors of plasmids also sell kits for the synthesis of probes from that plasmid. The kits come with specific instructions and the appropriate enzymes and reagents. Sometimes cDNAs are in plasmids that do not permit cRNA synthesis, so a procedure for inserting cDNA into a vector appropriate for making RNA probes is included. The rest of this chapter will be devoted to techniques for the isolation of RNA from tissue and quantitation of specific mRNAs.
II. Some General Rules
The single most important point to remember is that good results can be obtained only if one is fastidious, particularly when working with RNA. Ribonucleases (RNases), enzymes that degrade RNA, are ubiquitous and can quickly ruin a sample. It is imperative that gloves always be worn when working with RNA, as skin is a good source of RNases. Disposable plastic labware should be used whenever possible, as glass tends to harbor RNases. RNases are very difficult to denature, but glassware can be used after it has been baked at 150°C for 4 hours. Distilled water used as is or to make solutions should be treated with diethylpyrocarbonate (DEPC), an inhibitor of RNase, and then autoclaved. The final concentration of DEPC should be 0.1%. The notable exception is solutions buffered with Tris, which shou...
Table of contents
- Cover
- Title Page
- Copyright Page
- Dedication
- Table of Contents
- Part I. Molecular Techniques
- Part II. Protein Identification
- Part III. Cellular Techniques
- Index