
- 367 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Nanobiotechnology and Nanobiosciences
About this book
This volume introduces, in a coherent and comprehensive fashion, the Pan Stanford Series on Nanobiotechnology by defining and reviewing the major sectors of nanobiotechnology and nanobiosciences with respect to the most recent developments. It covers the basic principles and main applications of nanobiotechnology as an emerging field at the frontiers of biotechnology and nanotechnology, with contributions from leading scientists active in their respective specialties.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
Chapter 1
Nanoscale Materials
This chapter overviews the present status of new materials by organic and biological nanotechnology and their applications with respect to the development of organic and biological nanotechnology defining Nanobiotechnology and Nanobiosciences capable to yield a significant scientific and technological progress.
Particular emphasis is placed on what has been accomplished in our laboratory in the last eight years, whereby the details on the supramolecular layer engineering and its application to industrial nanotechnology can be found in recent complete reviews (Nicolini et al., 2001; 2005; Nicolini and Pechkova, 2006). Material technology has changed our lives within only a few decades.’ Sand, the starting material, has been turned into a versatile “high-tech” product. Techniques have been developed for the production of silicon wafers and for the modification of this raw material into the final “high-tech” product -miniature electronic logic functions. Research now focuses on the molecular manufacturing (Nicolini, 1996c), from the microstructures to the nanostructures for a new generation of nanomaterials. The rapid increase in our knowledge of the function of both biological and organic materials has also directed interest into this area. The functionality, efficiency, and flexibility of biomaterials (see paragraph 1.1) are impressive; they are beyond the capabilities of synthetic chemistry already quite powerful (see paragraph 1.2).
The development of gene-technological methods has opened the way to the controlled modifications of proteins, which appear already in position to allow the production of newly designed biomaterials. Several molecules of native organic and biological origin being investigated in the last years are here summarized to exemplify potentially useful nanomaterials. They can be divided into two major classes, namely those produced via biotechnology and those produced via chemical synthesis.
1.1 Produced Via LB Technology
1.1.1 Cells
Several well-established cell culture systems, like mammalian CHO-Kl, H9c2 or HeLa cells or microorganisms, are utilized in different context for various applications (Adami et al., 1992, 1995a; Hafemann et al., 1988; Garibaldi et al., 2006, Spera and Nicolini, 2007).
Chineese hamster ovary fibroblasts (CHO clone Kl) fior Nanoproteomics, as supplied by American Type Culture Collection, Rockville, MD, is a stable hypodiploid cell line derived by spontaneous transformation from a fibroblast culture (Misteli, 2001). Cells were cultured in F12 medium supplemented with 10% fetal calf serum and 0.2% gentamycin at 37 °C in 5% CO2 atmosphere. To reverse transform the cells they were treated with a solution 103 M dibutyryl cyclic 3’,5’ monophosphate adenosine sodium salt (Bt2cAMP, Sigma Chemical Co., St. Louis, MO) that was added to the normal culture medium for six hours before the analysis. It has been shown that single cells of CHO-Kl in the native state grow equally well on plastic surfaces or in suspension (Figure 1.1). In the presence of reverse transformation conditions, however, excellent growth is still achieved on the plastic surface but no growth whatever occurs in suspension (Hsie and Puck, 1971) as monitored by a phase contrast microscope (Wilovert, Wesco).
Primary cultures of rat hepatocytes for Nanosensors were also used for experiments with biosensing potentiometric systems because they are easy to obtain and, like all hepatocytes, they maintain, in the first hours in vitro, their metabolic skills practically unchanged with respect to the in vivo situation (Nicolini et al., 1995a). Hepatocytes were isolated from liver of Sprague-Dawley albino rats (200-250 g) by in situ collagenase perfusion according to Williams (Williams, 1977).
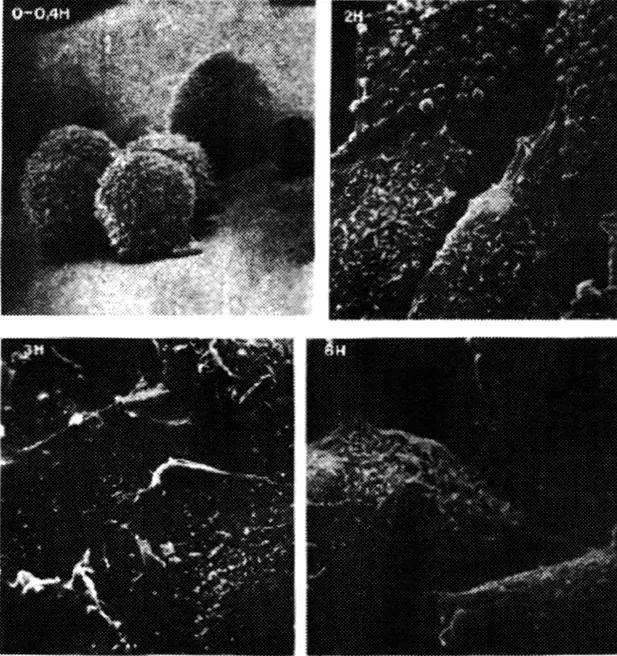
Figure 1.1 Chinese hamster ovary fibroblasts (CHO clone Kl). Electronic micrographs of CHO-Kl cells at t equal O hour (mitosis), 2 hours (Gl phase), 3 hours (S phase) and 6 (G2 phase) hours after mitosis. (Nicolini and Rigo, Biofisiche e Tecnologie Biomediche, Zanichelli Editore, 1992).
Cardiac muscle cells for Drug Delivery via Carbon Nanotubes are a rat heart cell line H9c2 (2-1), obtained from American Type Culture Collection (Rockville, MD) at passage 16. Cells at passages from 22 to 24 were used for the experiments. H9c2 cells were cultured in Dulbecco’s modified Eagle’s medium with 4 mM L-glutamine adjusted to contain 1.5 g/1 sodium bicarbonate, 4.5 g/1 glucose, 1.0 mM sodium pyruvate, 10% FBS, with 50 units penicillin/ml, 100 mg streptomycin/ml at 37°C in 5% CO2, and the medium was changed every 2-3 days. Subconfluent cells were detached with trypsin and seeded 24 hours before treatment at a density of 25000-30000 cells/cm2 in 100 mm petri dishes. Cells were seeded in Petri dishes and let to grow for 24 hours before treatments. While untreated control cells were administered the complete medium without nanotubes, sonicated in parallel with Single Wall NanoTube (SWNT) suspension, treated cells were administered complete medium with suspended 0.2 mg/ml SWNT for serial time steps. A positive control was set to induce cell damage and death by treating H9c2 with 200 μM and 1 mM hydrogen peroxide. The experiments were performed in triplicate. Cardiomyocytes treated or not with SWNT were evaluated at relevant time points by light microscopy to assess cell proliferation and viability. Digital photomicrographs were recorded and the number of viable cells computed. Myocytes were incubated for 24, 48 and 72 hours at standard culture conditions to determine the viability following treatments. The number of viable cells was determined with trypan blue exclusion. In brief, cell monolayers were rinsed twice with PBS and resuspended with trypsin and EDTA. The cells were immediately stained with 0.4% trypan blue, and the number of viable cells was determined using a hemocytometer under a light microscope. In order to assessment apoptosis the cells were labeled with annexin V-FITC and propidium iodide, and fluorescent positive cells were detected by flow cytometry. Cellular fluorescence was determined by a flow cytometry apparatus (FACS-SCAN, Becton-Dickinson, Franklin Lakes, NJ, USA). Flow cytometric analysis was performed on a minimum of 1 × 104 unfixed cells per sample. Cardiomyocytes were trypsinized with trypsin-EDTA, resuspended in PBS, and loaded with 10 μM propidium iodide (PI; Sigma-Aldrich) and 1 μM annexin V-FITC (Sigma Aldrich) at 4°C for 10 min. Apoptosis was identified as cells with low forward scatter (FSC) on side scatter (SSC)/FSC dot plots, PI dim staining on FSC/PI dot plots, and annexin V-positive and Pi-negative staining on annexin V/PI dot plots. Fluorescence probes were excited with an 488 nm argon ion laser. The emission fluorescence was monitored at 525 nm for annexin V-FITC and 620 nm for PI. Data were analyzed with the Cell Quest software package (Becton-Dickinson).
Human T lymphocytes for Nanogenomics, as previously shown (Nicolini et al., 2006a) are obtained by a sample of heparinized peripheral blood specimens diluted 1:1 with sterile phosohate buffer saline (PBS, pH 7.2). The sample was centrifiiged on a Ficoll-Hypaque gradient (specific gravity 1.080) at 1500 rpm for 40 min at room temperature without brake (Abraham et al., 1980). The lymphocytes rich interface was collected and washed twice with PBS, and the number of cells counted in presence of trypan blue. The 2 × 106 ceils were seeded into each well containing 2ml of RPMI 1640 culture medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin 100 unit/mL and streptomicin 50 pg/mL. The cells has then been incubated at 37 °C in a humid chamber with 5% of CO2 for 24, 48, 72 hours. The cells have been then collected and counted and then it has been proceeded to RNA extraction.
1.1.2 Proteins
Recombinant protein expression and purification is the single most important prerequisite for the effective engineering of nanostructured protein-based materials.
Typically the cDNA encoding the mature form of the protein being immobilized is subcloned into the proper expression vector, highly expressed in Escherichia coli and the expressed protein is typically purified by affinity chromatography (Pernecky and Coon, 1996; Ghisellini et al., 2004; Wada et al., 1991; Amann et al., 1988) and evaluated spectrophotometrically using the appropriate extinction coefficient at the appropriate wavelength.
In the case of overall cell proteins extraction, as required in Mass Spectrometry at the nanoscale, a protein extraction kit (Subcellular Proteome Extraction Kit, Calbiochem) is typically chosen for the total protein extraction from cells (Spera and Nicolini, 2007). It is designed for extraction of cellular proteins from adherent and suspension-grown cells according to their subcellular localization. For the sequential extraction of the cell content, the kit takes advantage of the differential solubility of certain subcellular compartments in special reagent mixtures. Upon extraction of culture cells, four partial proteomes of the cells are obtained:
- cytosolic proteins,
- membrane and membrane organelle proteins,
- nuclear proteins,
- cytoskeleton proteins
The kit contains four extraction buffers, a protease inhibitor cocktail to prevent protein degradation and a nuclease (Merck) to achieve an efficient removal of contaminating nucleic acids. The sub-cellular extraction was performed according to the protocol for freshly prepared adherent culture cells. We performed extraction from 106 CHO-Kl cells in logarithmic phase at 80% confluence (Spera and Nicolini, 2007). To monitor the extraction procedure, morphological changes of the cells were examined by a phase contrast microscope (Wilovert, Wesco). The protein fractions were stored at -80°C until the Mass Spectrometry analysis. The amount of total proteins recovered from each extraction step has been evaluated by Bradford assay (Tjio and Puck, 1958), using bovine serum albumin as standard.
1.1.2.1 Light sensitives
Photosynthetic reaction centers from Rhodobacter sphaeroides and bacteriorhodopsin (bR) from purple membrane (PM) have been used for their unique optoelectronic properties and for their capability of providing light-induced proton and electron pumping.
Once assembled they display extremely high thermal and temporal stability (Nicolini 1997a, 1998a,b, Nicolini et al., 1999). These features make them two of the most promising biological molecules for developing devices (Nicolini 1996a, Nicolini et al., 2005). RC proteins can conduct electrons in only one direction in presence of a suitable light source (photoconduction) or of an external electric field (conduction). Both these conductive processes are due to the tunnelling of a single electron into the structure of the protein.
In the case of bR the situation is different (Figure 1.2). BR is the main part of purple membranes (about 80%) and is already close packed in it. It is difficult to extract bR in the form of individual molecules, for they are very unstable (Shen et al., 1993). The modification of the retinal protein bacteriorhodopsin gave birth to the generation of new versatile media for optical processing (Zeisel and Hampp, 1992). Gene technology as the key technique in this new direction of nanomaterials research may be the counterpart to photolithography in semiconductor technology. The design of complex molecular functions is a goal extremely difficult to achieve wit...
Table of contents
- Cover
- Half Title
- Title Page
- Copyright Page
- Dedication Page
- Preface
- Table of Contents
- 1. Nanoscale Materials
- 2. Nanoscale Probes
- 3. Nanoscale Applications in Health and Science
- 4. Nanoscale Applications in Industry and Energy Compatible with Environment
- Bibliography
- Index
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access Nanobiotechnology and Nanobiosciences by Claudio Nicolini in PDF and/or ePUB format, as well as other popular books in Medicine & Biotechnology in Medicine. We have over one million books available in our catalogue for you to explore.