
- 384 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Practical Bioinformatics
About this book
Practical Bioinformatics is specifically designed for biology majors, with a heavy emphasis on the steps required to perform bioinformatics analysis to answer biological questions. It is written for courses that have a practical, hands-on element and contains many exercises (for example, database searches, protein analysis, data interpretation) to
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
CHAPTER 1
Introduction to Bioinformatics and Sequence Analysis
Key concepts
• The scope of bioinformatics
• The origins and growth of DNA databasess
• Evidence of evolution from bioinformatics
• Example sequence analysis and displays using human Factor IX
1.1 INTRODUCTION
We are witnessing a revolution in biomedical research. Although it has been clear for decades that exploring the genetics of biological systems was crucial to understanding them, it was far too expensive and complex to consider obtaining genetic sequences for that exploration. But now, acquiring genetic sequences is affordable and simple, and data are being generated at unprecedented rates. The heart of understanding all this sequence lies in bioinformatics sequence analysis, and this book serves as an introduction to this powerful study of DNA, RNA, and protein sequence.
Bioinformatics concerns the generation, visualization, analysis, storage, and retrieval of large quantities of biological information. The generation of biomedical data, including DNA sequence, in its raw form does not involve bioinformatics skills. But in order for that sequence to be usable, it must be analyzed, annotated, and reformatted to be suitable for databases. These are all bioinformatics activities. Many of these activities can be automated, but their development and support come from someone with skills or experience in bioinformatics.
Once the data have been made available, how do you analyze the data? Is there text like DNA and protein sequence files? If yes, it should be presented in a way to allow interpretation or easy input into programs for analysis. Or is there so much information that data are represented graphically? This form of data reduction is quite powerful and without it we would be staring at pages and pages of sequence without, literally, seeing the big picture.
Some analysis is manual, ranging from looking at the individual nucleotides or amino acids, to submitting sequence to a program that transforms the sequence into another form. This could include the location of features such as functional domains, modification sites, and coding regions. Often, analysis includes the searching of databases for the purposes of comparison or discovery, and this will be the primary activity for a number of chapters. Much of the content of this book is concerned with analysis.

In the early years of GenBank, if you wanted access to the database you ordered a handful of floppy disks that were delivered in the mail.
Storage is usually not a responsibility of those who will analyze the sequences. However, the creation of properly structured databases or storage forms so data can be queried and retrieved is essential for the analysts to do their work. Sequence files and other forms of data can be decades old or just created yesterday. But unless you can retrieve them easily, the value decreases quickly. “Easily” is not just describing the speed of the computers and connections delivering the information to you, although this can be extremely important. It also includes the steps to access and query the stored data. The ideal approach is often a Web form with easily understood options, online help, and results pages rich with hypertext. Bioinformatics was one of the first areas of science to embrace the Web as a vehicle for disseminating information and we’ll be using many Web pages in this book.
Finally, bioinformatics activities often involve large quantities of data. Even if you are focusing on a single gene, you still may have mountains of data that are connected to this single sequence. With a good database or software tool, you may only be aware of the quantities yet not overwhelmed with details that don’t interest you. Still, it can’t be emphasized enough that one of the biggest challenges facing the field of bioinformatics is the absolute deluge of information and how to generate, visualize, analyze, store, and retrieve these data.
1.2 THE GROWTH OF GenBank
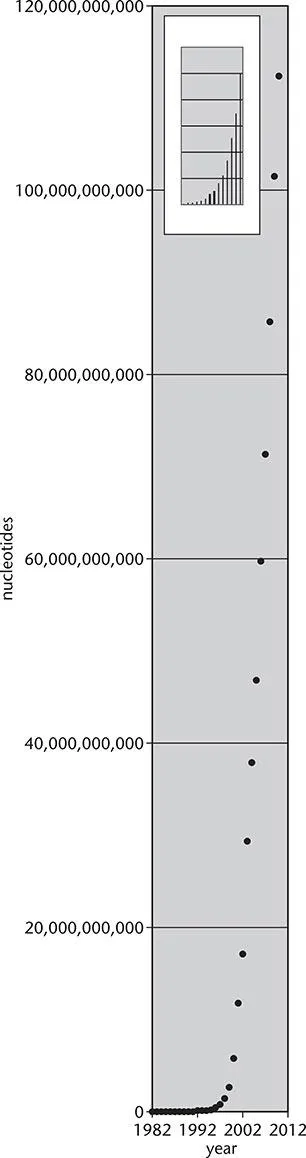
How much data are we talking about? One way to answer this is to describe the amount of DNA sequence data in public databases. GenBank is a huge repository run by the US National Center for Biotechnology Information (NCBI). The inset in Figure 1.1 shows the steady growth in the early years of GenBank but the rate of growth has been rapid since then. As of early 2011, there are over 126 billion nucleotides in this standard division of GenBank from over 380,000 organisms. If this were not impressive enough, there are an additional 91 billion nucleotides in the whole genome shotgun (a type of sequencing) division, the section of GenBank dedicated to unfinished large sequencing efforts. If a DNA sequence is considered “public information,” it is deposited in GenBank, the DNA Data Bank of Japan (DDBJ), or the database of the European Bioinformatics Institute (EBI). The contents of these three databases are synchronized. In terms of disk space, the database is over 500 Gigabytes in size.
1.3 DATA, DATA, EVERYWHERE
Where are all the data coming from? The quick answer is everywhere! In recent years there has been a dramatic drop in prices and rapid advances in both sequencing technology and computing power. What was once too time-consuming and expensive is now very possible and affordable; biological sequence generation is now commonplace. A major driver for the advances being realized today is the Human Genome Project. Even though the completion of the sequencing of the human genome was announced in 2001, the analysis of the data is ongoing and will take many years. These advances had to be coupled with dramatic improvements in computers and the drop in cost for processing power, memory, and storage. Of course, the Human Genome Project and all the spin-offs are only possible because of simultaneous advances in bioinformatics.
This intersection of sequencing technologies, computational power, and advances in bioinformatics has made DNA sequencing quite routine and paved the way for many bold and ambitious projects. Projects now come from scientists in numerous fields of biology, medicine, agriculture, ecology, history, energy, and forensics, just to name a few. Here are some prominent examples.

According to Eric Lander, director of the Genome Biology Program at the Broad Institute, it now costs about $20 to sequence the Escherichia coli genome, sequencing each of the 4.7 million nucleotides twenty times to ensure accuracy.
• The 1000 Genomes Project (www.1000genomes.org). An effort to sequence the genomes of 1000 people to identify genetic variants that affect 1% of the human population. In addition to providing insights to genetic disorders and health risks, the history of human migrations is being revealed. In recent years, people have proposed that the number of human genomes to sequence for this project grow to be 10,000 or higher.
• The 1001 Arabidopsis thaliana Genomes Project (www.1001genomes.org). Arabidopsis is a widely used plant model due to its habitat diversity, genetics, and ease of manipulation. This genome project aims to study the genomes of 1001 strains that differ in phenotype including adaptation to growth in a wide variety of conditions. Project scientists and those in the Arabidopsis community are able to grow huge numbers of genetically identical plants and can vary the environment at will to challenge and observe the underlying genetic elements which define these strains.
• The Genome 10K Project (http://genome10k.soe.ucsc.edu). An effort to sequence the genomes of 10,000 vertebrate species, one from every genus. Along with all the other genomes sequenced, this project will make a tremendous impact on understanding the relationship between organisms. We can only guess what will be discovered from these animals, having so much in common with us but with such diverse physiologies and phenotypes, and occupying such a wide range of habitats.
• The i5k Initiative (www.arthropodgenomes.org/wiki/i5K). An effort to sequence the genomes of 5000 insects and arthropods. Many insects are either pests, carriers of disease, or beneficial to agriculture and man. More knowledge of their biochemical pathways will surely result in new avenues of control, utilization, and fascination.
• Metagenomics. This is a broad term covering the sequencing of DNA samples from the environment as well as from biomedical sources. For example, sequencing has led to the identification of the hundreds of bacterial species inhabiting our skin, mouth, and digestive system. The populations that live on and within us vary with our health state and are clearly linked to our physiology (as we are to theirs). The NCBI lists almost 350 metagenomics projects (www.ncbi.nlm.nih.gov/genomes/lenvs.cgi) that are either at the beginning stages or completed. These projects each generate anywhere from thousands to millions of sequences.
• Cancer Genome Atlas. This is a massive project (http://cancergenome.nih.gov) where thousands of specimens from all the major cancer types and their matched normal controls will have their RNAs and many of their genes sequenced.
• EST generation. ESTs (expressed sequence tags) are small samples of transcribed genes and a quick avenue for discovering the genes expressed in tissues or organisms. Clones are generated and sequenced by the thousands. There are at least 72 million EST sequences in GenBank.
• The Barcode of Life (www.barcodeoflife.org). Distinguishing closely related species is often difficult, even for taxonomy experts. For example, there are approximately 11,000 species of ants. How can you easily tell them apart? The Barcode of Life project aims to identify a DNA “signature” for each species in the world using a 648 base pair sequence of the cytochrome c oxidase 1 gene. The five-year goal is to have sequences from 500,000 species. Nice examples of consumer use of this information include the identification of illegal fishing of endangered species and illegal logging activities.
• The NCBI lists over 1700 eukaryotic genome sequencing projects (www.ncbi.nlm.nih.gov/genomes/leuks.cgi), over 11,000 microbial genome projects (www.ncbi.nlm.nih.gov/genomes/lproks.cgi), and over 3100 viral genomes.
There are also private sequencing efforts where the data are not always released to the public yet the parties acquiring the data still have to cope with the huge amount of sequence generated by these projects.
...Table of contents
- Cover
- Half Title
- Dedication
- Title Page
- Copyright Page
- Preface
- Acknowledgments
- Table of Contents
- Chapter 1 Introduction to Bioinformatics and Sequence Analysis
- Chapter 2 Introduction to Internet Resources
- Chapter 3 Introduction to the BLAST Suite and BLASTN
- Chapter 4 Protein BLAST: BLASTP
- Chapter 5 Cross-Molecular Searches: BLASTX and TBLASTN
- Chapter 6 Advanced Topics in BLAST
- Chapter 7 Bioinformatics Tools for the Laboratory
- Chapter 8 Protein Analysis
- Chapter 9 Explorations of Short Nucleotide Sequences
- Chapter 10 MicroRNAs and Pathway Analysis
- Chapter 11 Multiple Sequence Alignments
- Chapter 12 Browsing the Genome
- Appendix 1 Formatting Your Report
- Appendix 2 Running NCBI BLAST in “batch” Mode
- Abbreviations
- Glossary
- Web Resources
- Index
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access Practical Bioinformatics by Michael Agostino in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.