
eBook - ePub
Quick Guideline for Computational Drug Design
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Quick Guideline for Computational Drug Design
About this book
Bioinformatics allows researchers to answer biological questions with advanced computational methods which involves the application of statistics and mathematical modeling. Structural bioinformatics enables the prediction and analysis of 3D structures of
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Quick Guideline for Computational Drug Design by Sheikh Arslan Sehgal,A. Hammad Mirza,Rana Adnan Tahir in PDF and/or ePUB format, as well as other popular books in Sciences physiques & Chimie. We have over one million books available in our catalogue for you to explore.
Information
3D Structure Prediction
Sheikh Arslan Sehgal, A. Hammad Mirza, Rana Adnan Tahir
Abstract
The proteins having similar sequences may have similar structure” is the philosophy of sequences prediction techniques. The 3D structures have structural and functional importance for drug designing. Homology modeling, threading and ab initio approaches are the computational techniques to predict the 3D structure of the proteins for further studies. In this chapter, we will precisely elaborate numerous tools of 3D structure prediction.
Keywords: ab initio, Homology modeling, I-Tasser, MODELLER, Structure prediction, Threading.
Motivation
3D structure of a protein is important for the sequences and functional analysis of a protein. Experimentally determined structures through X-ray crystallography and Nuclear Magnetic Resonance (NMR) techniques provide detailed information related to the dynamics of the protein interactions with ligands, receptors, interacting proteins, and with its environment. There are 35,914 structures of human sequences available in PDB but still, usually the protein of interest may not have its structure solved through experimental techniques. In such conditions, it becomes necessary to have opted different methodologies for structural analyses. Computational methods are providing alternative ways to solve this problem by utilizing the structure prediction techniques. This chapter is aimed to introduce different desktop and web-based structure prediction methods. Many tools will be discussed and each tool implementing a different algorithm to model the protein structures. It will help the researchers and also the beginners to understand 3D structure prediction and can also compare the results according to the behavior of the protein of interest.
“The proteins having similar sequences may have similar structures” is the philosophy of structure prediction techniques. Due to unavailability of resources and time trade-offs, it is not possible to find the structure of proteins through X-ray crystallography and NMR techniques.
Structure prediction tools help to solve the 3D structure of proteins based on similarity (homology) of template protein structure with query protein (protein of interest). All of the structure prediction algorithms try to first align sequences of the protein of interest with the template(s), so first most important step in structure prediction is alignment. In this Chapter, we will follow the methodology of section 04 mentioned in Fig. (3).
Structure Prediction Tools
Protein structure prediction is an effort to predict the 3D structure of a protein from its amino acid sequence. The primary sequence also used to predict the folding and its secondary, tertiary, and quaternary structure (Mount et al., 2001).
A large number of protein structure predictions are very expensive and time consuming by X-Ray crystallography and NMR methods (Sehgal et al., 2014). Numerous computational approaches and tools for 3D structure prediction have been developed. It is critical that the biological community and beginners are aware of such tools and is able to interpret their results in an informed way. The elaborative methodology may provide a guideline to predict the 3D structure by different approaches and also interpret the results effectively.
Phyre2
Phyre2 is a structure prediction non-commercial tool and is able to regularly generate the reliable protein models (Lawrence et al., 2011).
Introduction
Phyre2 server predicts the 3D structure of target protein sequence by using homology modeling approach. It has been ranked amongst one of the best systems of its kind for the last four years as judged by the biannual Critical Assessment of Structure Prediction (CASP) meetings (Lawrence et al., 2011).
Brief Instructions
Following instructions should be followed in order to predict the 3D structure of the respective protein.
- Open the web browser
- Go to Phyre2 homepage (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)
- An interface will open where the amino acid sequence of the respective protein is uploaded e.g. RASSF2
- Provide the institutional e-mail ID ([email protected])
- Results will be sent through e-mail depending upon the sequence (query) length and number of protein sequences in queues
Requirements
Input
The input required for Phyre2 is the FASTA sequence of target protein retrieved from UniProt.
Sequence Submission
The FASTA sequence of RASSF2 submitted to phrex2 (Fig. 8).
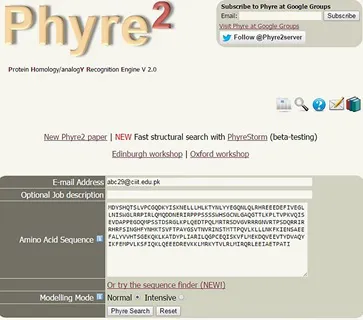
RASSF2 Sequence submission to Phyre2.
Click on the “Phyre search” option and the generated 3D structure will send to the mentioned mail ID.
Results Interpretation (Results Screen)
The detailed information about the target protein (RASSF2) can be retrieved by opening the Phyre2 link sent to the user by e-mail containing results. The results screen of Phyre2 is divided into following main sections:
Secondary Structure and Disor...
Table of contents
- Welcome
- Table of Contents
- Title
- BENTHAM SCIENCE PUBLISHERS LTD.
- FOREWORD
- PREFACE
- Introduction to Structural Bioinformatics
- Protein Primary Sequence Analysis
- Secondary Structure Analyses
- 3D Structure Prediction
- Structure Evaluation
- Visualization of Predicted Structure
- Molecular Docking Studies