
- 436 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Advanced Diffusion Encoding Methods in MRI
About this book
The medical MRI community is by far the largest user of diffusion NMR techniques and this book captures the current surge of methods and provides a primary source to aid adoption in this field.
There is a trend to adapting the more advanced diffusion encoding sequences developed by NMR researchers within the fields of porous media, chemical engineering, and colloid science to medical research. Recently published papers indicate great potential for improved diagnosis of the numerous pathological conditions associated with changes of tissue microstructure that are invisible to conventional diffusion MRI. This book disseminates these recent developments to the wider community of MRI researchers and clinicians. The chapters cover the theoretical basis, hardware and pulse sequences, data analysis and validation, and recent applications aimed at promoting further growth in the field.
This is a fast moving field and chapters are written by key MRI scientists that have contributed to the successful translation of the advanced diffusion NMR methods to the context of medical MRI, from global locations.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
1.1 Introduction
1.2 A Molecular Perspective on Water Diffusion in Biological Tissues
1.2.1 Biomembranes
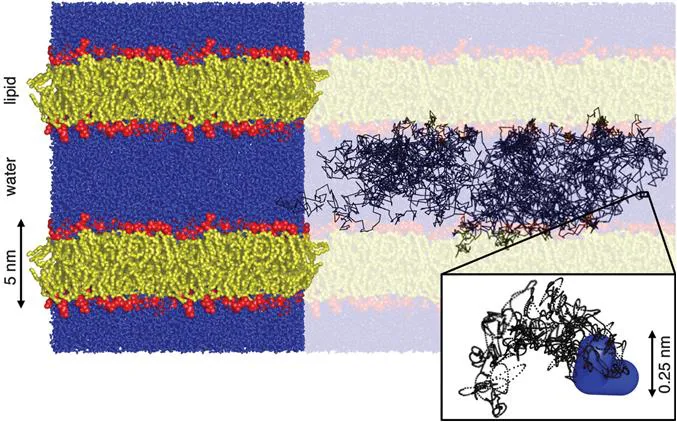
1.2.2 Macromolecules
Table of contents
- Cover
- Title
- Copyright
- Preface
- Contents
- Chapter 1 Translational Motion of Water in Biological Tissues – A Brief Primer
- Chapter 2 Diffusion Encoding with General Gradient Waveforms
- Chapter 3 Diffusion Anisotropy and Tensor-valued Encoding
- Chapter 4 Restricted Diffusion and Spectral Content of the Gradient Waveforms
- Chapter 5 Separating Flow from Diffusion Using Velocity-compensated Diffusion Encoding
- Chapter 6 Disentangling Intercompartment Exchange from Restricted Diffusion
- Chapter 7 Estimating Chemical and Microstructural Heterogeneity by Correlating Relaxation and Diffusion
- Chapter 8 Hardware for Generating Modulated Gradient Waveforms with High Precision
- Chapter 9 Pulse Sequences Combining Advanced Diffusion Encoding and Image Read-out
- Chapter 10 Nonparametric Inversion of Relaxation and Diffusion Correlation Data
- Chapter 11 Model-based Analysis of Advanced Diffusion Data
- Chapter 12 Phantoms for Validating Advanced Diffusion Sequences
- Chapter 13 Advanced Histology for Validation of Advanced Diffusion MRI
- Chapter 14 Clinical Research with Advanced Diffusion Encoding Methods in MRI
- Subject Index
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app