
- 664 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Molecular Pathomechanisms and New Trends in Drug Research
About this book
Knowledge of the basic mechanisms of human disease is essential for any student or professional engaged in drug research and development. Functional gene analysis (genomics), protein analysis (proteomics), and other molecular biological techniques have made it possible to understand these cellular processes, opening up exciting opportunities for no
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Molecular Pathomechanisms and New Trends in Drug Research by Gyorgy Keri,Istvan Toth in PDF and/or ePUB format, as well as other popular books in Medicina & Farmacología. We have over one million books available in our catalogue for you to explore.
Information
Part 1
Introduction
Chapter 1
Introduction
A breakthrough in modern drug research
György Kéri and István Toth
In recent years dramatic developments in molecular biology, pathobiochemistry and bioinformatics have greatly increased our understanding of the molecular pathomechanisms of various diseases. With the help of functional gene analysis (genomics), functional protein analysis (proteomics), molecular diagnostics, and other molecular biological techniques it has become possible to understand in more detail the molecular mechanisms which contribute to complex pathological states, opening up novel therapeutic possibilities.
Molecular pathomechanisms of diseases are usually very complex; their complete understanding still demands a lot of effort. The first question is how do we define the term “pathomechanism”? The greek word “patho” means disease, so pathomechanism means disease-mechanism, while in molecular terms we must clarify the differences in the molecular mechanisms of the normal and the pathological state. This question will lead us to the more philosophical one: what is the normal state, when do we function properly? Without getting too philosophical it is worthwhile reminding ourselves of an old Chinese saying:
The unity of spirit, soul, body and mindWill make you happy and shine
In other words, the normal state is when we function properly, think properly, feel properly and act properly, or in a more simple way when we have peace of mind because we have a meaningful life, we know what we want and we do it.
In contrast, in a pathological state due to physical, mental or emotional problems (or a mixture of all of them), we do not function properly, and in most cases these problems cannot be localized to a particular organ or area of the body, as the pathological state usually means a systemic problem. If certain parts of the organism do not fulfill their task or duty, this influences or inhibits the proper functioning of the whole organism, resulting in system mal-function. For example, cancer is the result of genomic changes, which usually develops from a single transformed cell (monoclonal origin). However whether such a transformed cell can become a fully fledged malignant tumor very much depends on the systemic response, on the intra- and intercellular (and may be even interpersonal) relationships and communication. This systemic view is also true for pathological states caused by external factors, for example for viral or bacterial diseases, where the pathogens take over and reprogram the communication channels of the host cell in order to serve the intruder optimally.
Recently, it has become evident that, in most cases, intraor intercellular communication disorders form the background to complex pathomechanisms. Thus modern drug research has focused on signal transduction therapy, with the primary task of understanding the relevant molecular pathomechanism.
The aim of this book is to present a relatively broad picture about molecular pathomechanisms and new trends in drug research, with a special emphasis on signal transduction problems and potential therapeutic strategies.
Many molecular pathomechanisms are the result of an intracellular or intercellular communication disorder, while a series of genomic changes can be the cause and the consequence of these communication disorders. In a healthy organism, normal cells fulfill their duties, do not send or receive false messages and are strongly controlled by the external messages of the communication network. On the other hand, for example, cancer cells generate a false, mimicked proliferation signal for themselves via oncogenes and other genomic changes.Whether this communication failure is the result of environmental factors and/or external messages (generating changes at the genomic level), or originates in the genetic program is still a question, and can be answered only on a case-by-case basis. However we have to consider that cells, like human beings, live in a well organized society and in a given ecosystem, which, to a certain extent, determines their receptivity and responsiveness as well as the systemic response for the various carcinogenic agents and effects. In other words, carcinogenic compounds can be carcinogenic in a given in vitro system or in a given organism, but the same agent can have different effects in different systems, depending, of course, on the extent of the effect. Clearly changes at the genomic level are critical steps during carcinogenesis, however the manifestation of these genomic changes and the system response depends very much on the communication state and responsibility of the system.
The processes of cellular growth and differentiation as well as the maintenance of specialized functions show a remarkable degree of coordination and it has been clearly demonstrated that this involves intercellular communication, rather then relying entirely on intracellular programing. In the pathological state, the normally interdependent system controls are uncoupled and certain cellular functions or malfunctions are stimulated in such a way as to result in further damagecausing signals, or often in the growth of the malfunctioning cells. Proliferation of infected, damaged or malfunctioning cells is very often a key factor in the generation of the pathological state, not only in cancer and infectious diseases but also in inflammation or autoimmune-related diseases like arteriosclerosis, arthritis, or certain inflammation-related neurodegenerative diseases. Inflammation has been found to be a determinatory pathological cause of many chronic diseases, where proliferation of immune cells due to false signaling turned out to be a critical factor in the generation of the disease.
In the above mentioned cases, disease manifestation starts when these malfunctioning or transformed cells diversify, and variants with altered properties arise in the population. To survive and function in a competitive environment, such variants must have selective growth and communicative properties and other competitive advantages over other cells. The surviving malfunctioning cells must have specific signal transduction pathways turned on, with which all the feedback effects and inhibitory actions of the microenvironmental parameters cannot interfere. Since communication disorders represent a major cause of pathological states and most of the recently identified validated target molecules of drug research are signal transduction related macromolecules, most of the pathomechanisms and drug research areas described in this book relate to signal transduction.
This book describes several general strategies and tactics and related results for molecular pathomechanism-based target selection and validation. This is an area where the most dramatic progress has occurred in recent years. A series of genomic and proteomic approaches together with bioinformatic tools have been developed and used to identify novel molecular pathomechanism-related drug discovery targets. Functional genomics (not only in the human genome project but also in model organisms, pathogens and experimental animals) and proteomics in conjunction with an informationintensive “knowledge base” approach for comparative genomics and proteomics has generated a large amount of data for pathological target selection and validation. Dominant negative mutants, antisense, ribozyme and antibody methods and other modern molecular biological techniques have been used successfully for molecular target validation. For example cDNA microarrays and two-dimensional gel electrophoresis unmask the expression of genes with unassigned or unexpected functions, while depletion of mRNA with ribozymes or neutralization of proteins with intracellular antibodies or small molecular inhibitors enable the investigators to select and validate relevant target molecules for certain diseases.
Most of the potential novel molecular targets for signal transduction therapy can be grouped into the following categories: growth factor-, hormone-, cytokine receptor targets, tyrosine kinases and serine/threonine kinase signal transduction pathway targets; cell cycle targets; apoptosis-related targets; extracellular matrix targets, immune cell receptor targets, angiogenesis and metastasis targets; and cell life-span targets. On the other hand, because of the recent emphasis on the systemic nature of cellular and pathological states, genome-wide expression monitoring and analysis of genomic networks has become a novel tool of functional genomics. This method is aimed at identifying groups of coregulated genes and discovering genes expressed differentially in distinct situations. Such a global (genome-wide) view of “gene function” in the regulation of the dynamic relationship between proliferation, differentiation and apoptosis can provide new insights into cellular homeostasis.
Computational biology and bioinformatics is becoming increasingly important in pathomechanism-based drug target selection and validation. An integrated “knowledge base” approach is required to understand the interrelationship between genetic information, transcription and translation, the existing phenotype and cellular function and how this is modified by environmental factors. Cell biology, molecular medicine and genomic technologies are inseparable for defining and validating new molecular targets.
Recently developed pharmacogenomic approaches provide additional research tools for modern drug research elucidating the genetic polymorphisms in drug-metabolizing enzymes, transporters, receptors, and other drug targets. These have been linked to interindividual differences in the efficacy and toxicity of many medications, thus providing novel target molecules and putting more emphasis on novel drug delivery and formulation approaches. Drug delivery has also become a key issue in modern drug research, not only to improve bioavailability of the drug molecules by utilizing passive and active transport systems, but to target the drug to its active site. In addition, rational drug design, combinatorial chemistry and high-throughput screening against purified molecular and cellular targets have emerged as powerful techniques for target oriented drug discovery.
Without an exhaustive description of all known molecular pathomechanisms, a series of representative diseases have been discussed in this book. The examples have been selected on the basis of pathological and epidemiological significance and on the basis of the present trends in modern drug research. The diseases discussed provide examples of signal transduction therapy and the systemic nature of the molecular pathomechanisms of the diseases, indicating that rate limiting steps or key signaling elements can become validated targets of modern drug research, reminding us of the saying:
The sea is in the drop and the drop is in the sea.
Part 2
Pathomechanisms and molecular target finding
Chapter 2
Drug discovery based on functional genomics
Target selection and validation
Gábor Mikala and István Vályi-Nagy
Discovery and development of therapeutically useful drugs requires the concentration of resources. Traditionally, models of human disease are utilized for drug screening purposes. Most frequently, these are animal or cell-based screening models representing a well-defined disease phenotype. Drug candidates are tested in these models in order to identify compounds that alter the disease phenotype. The majority of the therapeutically available drugs used in the Western world were developed along these “traditional” avenues. The main advantage of this screening method is that it uses a full biological system, and compounds passing this test are highly likely to be useful as therapeutic agents. Parallel to this, serious toxicity can also be ruled out. The major disadvantage of this approach rests in its inability to pinpoint target molecules, therefore, one cannot predict and dissect mechanisms of action and side effects. As a consequence, there are still a number of widely used drugs with obscure molecular mechanism of action.
Another “traditional” approach to drug selection has been aided by developments in protein purification and in molecular biology. Here, a molecular target of drug action is identified first, a molecule that is predicted to have a rate-limiting role in the disease development or phenotype. Once a target molecule is defined, potent and effective small molecules may be identified that have a well-defined mechanism of action and in vitro activity. Unfortunately, toxicity, metabolism and poor bioavailability are problems frequently encountered in the process of development of candidates with this approach. Moreover, since this procedure starts with a target molecule that plays a pivotal role in the disease process, correct identification and proper evaluation of the target “receptor” is essential for eventual success. Clearly, both “canonical” methods of drug screening have led to the development of many life-saving and life-improving medicines, however, they are also inherently responsible for the very high and costly attrition rate in drug development. Should it be possible to screen a large number of candidate “receptors” and drug molecules against one another, cost containment for drug development may be found.
The advent of the genomics era and development of numerous new screening procedures have the potential to provide the raw material for a revolution in pharmaceutical research. The human (and human pathogenic organism) genome projects have given rise to a tremendous number of target genes for screening, and combinatorial chemistry has the potential to provide countless compounds to test against them. Unfortunately, the complexity of both approaches is such that homing in on the right drug-receptor pair remains quite a challenge. In this chapter the authors attempt to introduce the reader to some of the revolutionary techniques that help to select and validate the molecular targets for drug development in the genomics era (Figure 2.1).
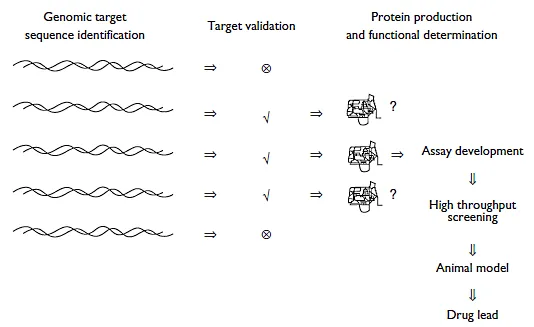
Figure 2.1 Classical drug discovery process based on functional genomics. In the genomic era much of the drug discovery process is still done in the traditional manner, target selection and validation (and frequently detailed functional determination) are placed upstream of assay development and drug screening. The major disadvantage of this time-tested approach is that it cannot be scaled up effectively to handle the large numbers of candidate drug target genes provided by genome projects.
2.1 Genomics-based drug target selection
The first step in genome-wide DNA sequence analysis has been the identification and tagging of each gene (expressed sequence tags – ESTs) in the human genome. At the same time, the genomes of several important microorganisms that are pathogenic to humans were sequenced. Both approaches provide a plethora of genetic information that is expected to deliver approximately 10,000 new therapeutic targets. Unfortunately, it has been difficult to pinpoint the most important drug-receptor candidates since more than three quarters of the proteins predicted to exist based exclusively on their corresponding cDNA sequence have no known function. The scientific fields of functional genomics and proteomics have developed out of necessity to handle this mass of data. Proteomics (in analogy to genomics that aims to provide sequence information on nucleic acids of a species, the genome) seeks to provide functional information for all proteins, the proteome. EST database mining, expression analysis and proteomics technologies are the most important new avenues in drug target discovery of the genomics era (Jones, DA et al. 1999, Johnston et al. 1999).
2.1.1 EST database mining based on homology searches
There are huge databases that contain EST information on the human genome. There are two main methods of utilizing these databases in the discovery process. First, it is possible to identify new members of previously known target protein families. This goal may be reached by simply searching in the databases for new sequences that possess structural features known to be characteristic for a certain class of proteins. In the simplest case this structural feature is a certain sequence to be found in the cDNA (by reverse translation). This approach has been particularly useful in identifying new ion channel subunits or proteins that regulate apoptosis or inflammation. New findings include isoforms of the much sought-after T-type calcium channel or novel cytokines related to tumor necrosis factor (TNF ). Searching of databases using a universally available Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) and the amino acid sequence of another calcium channel or TNF, respectively, helped to identify these novel molecules. Follow-up cloning and expression studies have confirmed that the identified targets are really the T-type calcium channel (and several distinct isoforms) or TNF-like proteins. At this point the “conventional” molecular-based drug discovery processes may take over and utilize the new protein as a drug screening target as in the case of the new ion channel. Alternatively, as in the case of the new TNF-analogues, though new pharmacological intervention points to inflammatory pathways may exist, more pre-drug-development research is needed to identify the correct targets among them for further drug discovery.
2.1.2 Differential tissue expression revealed by EST databases
The other route available for use of EST databases in the selection of possible targets is through the use of differential gene expression as revealed by tissue specific EST databases. EST databases currently encompass several different human tissues. Since the ideal drug target is expressed only in a single tissue or cell type – i.e. the diseased one to be modified by the drug under development – selection of a target molecule specifically expressed in that particular tissue is especially promising for further drug development. Theoretically, these tissue-restricted genes provide drug targets with great specificity and a reduced number of potential side effects. An example for this type of target selection approach is the identification of cathepsin K (an osteoclast-specific protease) as a promising drug target (Drake et al. 1996). Cathepsin K was discovered in a human osteoclast EST library through homology matches to known cathepsin, and further experimentation proved the molecule to be exclusively expressed in osteoclasts. Since pronounced bone matrix protein degradation was thought to be characteristic to the bone disease osteoporosis, inhibitors for cathepsin K were produced and tested in in vitro and in vivo models of bone resorption.
2.1.3 Expression analysis of diseased tissues for target identification
Database minings through homology searches and differential tissue expression are powerful tools for target selection, however, they have one crucial drawback. They could only be used effectively for selection of targets that have certain features that place them into a category of homologous proteins with known function. Unfortunately, most of the possible targets have no known structural relative. On the other hand, in certain settings it may not be crucial to have a known homologue of a target gene product.
Expression microarrays – or DNA-chips – are novel molecular tools that allow comprehensive analysis of RNA expression in different cells or tissues (e.g. healthy versus diseased) (Schena et al. 1998, Harrington et al. 2000). Though the methodology is detailed elsewhere in this book, it is useful to outline the essence of this new tool here. Expression microarray analysis involves quantitative hybridization of a large set of DNA probes (currently in the or...
Table of contents
- Cover Page
- Title Page
- Copyright Page
- Contributors
- Acknowledgments
- Aims and scope
- Part 1: Introduction
- Part 2: Pathomechanisms and molecular target finding
- Part 3: Common pathway and general mechanism
- Part 4: Drug discovery
- Part 5: Molecular pathomechanism of cancer
- Part 6: Infectious diseases
- Part 7: Diseases of the central and the peripheral nervous system
- Part 8: Cardiovascular diseases
- Part 9: Endocrinal and gastrointestinal disorders
- Part 10: Drug applications