
- 516 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Epigenetic Technological Applications
About this book
Epigenetic Technological Applications is a compilation of state-of-the-art technologies involved in epigenetic research. Epigenetics is an exciting new field of biology research, and many technologies are invented and developed specifically for epigenetics study. With chapters covering the latest developments in crystallography, computational modeling, the uses of histones, and more, Epigenetic Technological Applications addresses the question of how these new ideas, procedures, and innovations can be applied to current epigenetics research, and how they can keep pushing discovery forward and beyond the epigenetic realm.- Discusses technologies that are critical for epigenetic research and application- Includes epigenetic applications for state-of-the-art technologies- Contains a global perspective on the future of epigenetics
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
The State of the Art of Epigenetic Technologies
Keywords
1.1 Epigenetics and Chromatin Function
1.2 Mechanisms of Epigenetic Regulation
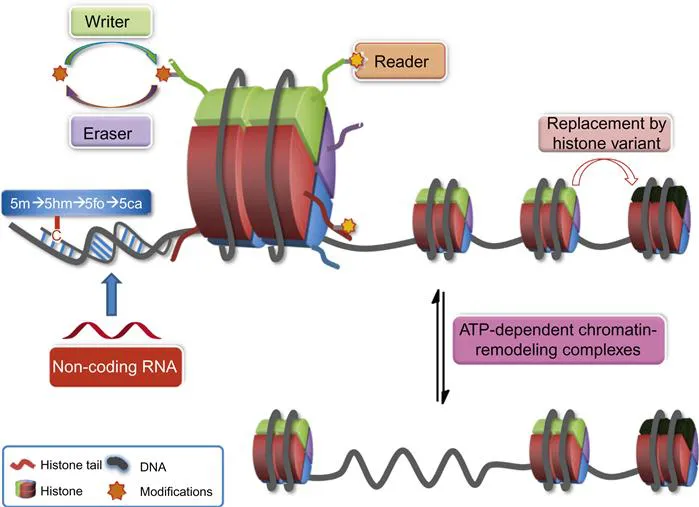
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- List of Contributors
- Foreword
- Chapter 1. The State of the Art of Epigenetic Technologies
- Chapter 2. Technologies for the Measurement and Mapping of Genomic 5-Methylcytosine and 5-Hydroxymethylcytosine
- Chapter 3. High-Throughput Sequencing-Based Mapping of Cytosine Modifications
- Chapter 4. Application of Mass Spectrometry in Translational Epigenetics
- Chapter 5. Techniques Analyzing Chromatin Modifications at Specific Single Loci
- Chapter 6. Comprehensive Analysis of Mammalian Linker-Histone Variants and Their Mutants
- Chapter 7. Crystallography-Based Mechanistic Insights into Epigenetic Regulation
- Chapter 8. Chemical and Genetic Approaches to Study Histone Modifications
- Chapter 9. Peptide Microarrays for Profiling of Epigenetic Targets
- Chapter 10. Current Methods for Methylome Profiling
- Chapter 11. Bioinformatics and Biostatistics in Mining Epigenetic Disease Markers and Targets
- Chapter 12. Computational Modeling to Elucidate Molecular Mechanisms of Epigenetic Memory
- Chapter 13. DNA Methyltransferase Inhibitors for Cancer Therapy
- Chapter 14. Histone Acetyltransferases: Enzymes, Assays, and Inhibitors
- Chapter 15. In Vitro Histone Deacetylase Activity Screening: Making a Case for Better Assays
- Chapter 16. Enzymatic Assays of Histone Methyltransferase Enzymes
- Chapter 17. Histone Methyltransferase Inhibitors for Cancer Therapy
- Chapter 18. Discovery of Histone Demethylase Inhibitors
- Chapter 19. Histone Demethylases: Background, Purification, and Detection
- Chapter 20. Animal Model Study of Epigenetic Inhibitors
- Index
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app