
- 268 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Metabolomics as a Tool in Nutrition Research
About this book
Metabolomics is a multidisciplinary science used to understand the ways in which nutrients from food are used in the body and how this can be optimised and targeted at specific nutritional needs. Metabolomics as a Tool in Nutrition Research provides a review of the uses of metabolomics in nutritional research. Chapters cover the most important aspects of the topic such as analysis techniques, bioinformatics and integration with other 'omic' sciences such as proteomics and genomics. The final chapters look at the impact of exercise on metabolomic profiles and future trends in metabolomics for nutrition research.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Metabolomics as a Tool in Nutrition Research by J-L Sebedio,L Brennan in PDF and/or ePUB format, as well as other popular books in Technology & Engineering & Nutrition, Dietics & Bariatrics. We have over one million books available in our catalogue for you to explore.
Information
Part One
Principles
1
Challenges in nutritional metabolomics
from experimental design to interpretation of data sets
M. Ferrara; J.-L. Sébédio Institut National de la Recherche Agronomique (INRA), Clermont-Ferrand, France
Abstract
High-throughput technologies (transcriptomics, proteomics, and metabolomics) that enable large-scale molecular measurements have begun to attract the attention of researchers in the medical, biological, and nutritional sciences because these tools can potentially characterise global alterations associated with disease conditions and nutrition, moving nutrition from a rather reductionist approach to a global one. This chapter describes the major challenges that scientists must face in order to develop metabolomics to its full potential, including experimental and analytical designs, analysis, and the biological interpretation of data sets.
Keywords
Metabolomics
Nutrition
Biomarkers
Bioinformatics
Structure elucidation
Analytical platforms
Statistics
Biological interpretation
Standardisation.
1.1 Introduction
Many studies have shown that diseases are often linked to poor nutrition (Aburto et al., 2013; Micha et al., 2013; Simopoulos, 2013; Wang et al., 2013). Nutrition research has identified the functions of essential nutrients, and researchers have established nutrition recommendations that allow the public to meet the needs of their bodies by optimising metabolic pathways. As an example, nutrition research led to the discovery of linoleic (18:2n− 6) and linolenic acids (18:3n− 3), which are fatty acids involved in essential functions that must be provided by food components because they cannot be synthesised by the body (Holman, 1996). However, considering interindividual variability in response to food components, an individual’s optimum physiological function must first be determined in order to optimise recommendations and achieve personalised nutrition (Zivkovic and German, 2009). Food science and nutrition not only focus on the relationship between diseases and macro- and micronutrient consumption, but these disciplines also attempt to identify bioactive molecules that may be present in the diet in low quantities for the purpose of understanding the protective effects that these molecules may have and their mechanisms of action. These non-essential bioactive compounds interact with a number of metabolic pathways and may modulate health status, either directly or indirectly, after being metabolised by the gut microbiota. Dietary polyphenols are a typical example of the importance that gut microbiota may play (Moco et al., 2012).
Consequently, identifying specific early-responding molecules in biofluids, such as blood and urine, is crucial for diagnosis and intervention, or process reversal (Reezi et al., 2008), because these substances may be connected to underlying metabolic dysfunctions. For this purpose, researchers must obtain accurate data on dietary intake in natural populations, which is a great challenge for epidemiological studies (Ismail et al., 2013). The recent development of high-throughput technologies (transcriptomics, proteomics, and metabolomics), which permit large-scale molecular measurements, has drawn the attention of scientists in the medical, biological, and nutritional fields because these tools offer the possibility of characterising global alterations associated with disease conditions, exposing overall nutritional activities and moving nutrition from a rather reductionist approach to a global one. The production and utilisation of metabolites is more directly connected to the phenotype exhibited by an organism than the presence of mRNAs or proteins (Figure 1.1).
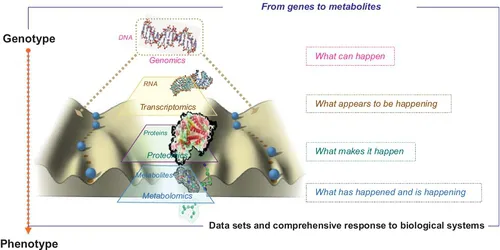
Figure 1.1 Metabolomics: bridging the genotype–phenotype gap. The “omic” technologies are used to obtain a more holistic view of how biological systems work and underpin the fields of functional genomics and systems biology. Genes encode mRNAs that encode proteins that, with environmental factors (e.g., diet), lead to the metabolite.
Among the “omic” technologies, the metabolomics has mainly grown out of the pharmaceutical and plant sciences. Yet, despite its relatively recent application to nutrition, this topic constitutes a growing domain of interest, considering the shift observed in dietary habits (German et al., 2002; Gibney et al., 2005). The rapidly expanding field of metabolomics has been driven by major advances in analytical tools, such as Nuclear Magnetic Resonance (NMR) (Eisenreich and Bacher, 2007) and mass spectrometry, and the corresponding hyphenated methods, such as high-performance liquid chromatography coupled with mass spectrometry (LC-MS) and gas–liquid chromatography coupled with mass spectrometry (GC-MS) (Dettmer et al., 2007; Pan and Raftery, 2007; Wilson et al., 2005). Advances in chemometric and bioinformatic technologies (Idle and Gonzalez, 2007; Moco et al., 2007; Wishart, 2007) also play an important role. Metabolomics (Fienh et al., 2000; Nicholson et al., 1999) can be described as a global analysis of small molecules of a biofluid (blood, urine, culture broth, cell extract, etc.), which are produced or modified as the result of a stimulus (nutritional intervention, environmental stressors, drug, etc.). At the moment the study of lipid molecules (lipidomics) is a major area in the metabolomics field (Morris and Watkins, 2005; see Chapter 3 for detailed discussion). Some chapters of this book will also discuss the use of metabolomics for discovering biomarkers in nutritional interventions (Brennan, 2013; Lodge, 2010), for predicting the risk of a health state involving into a disease state, and for monitoring food/nutrient biomarker consumption (Hedrick et al., 2012; Pujos-Guillot et al., 2013).
A typical metabolomic experiment (Figure 1.2) includes six elements: (1) design of the experiment and preparation of the sample; (2) analysis of biofluids or tissues via LC-MS, NMR, GC-MS, or a multiplatform approach to maximise metabolite coverage; (3) extraction and treatment of the data set; (4) statistical analyses; (5) identification of key or discriminatory metabolites; and (6) biological interpretation of the data. Data acquisition and retrieval may be achieved by a targeted analysis during which specific pathways are analysed or an untargeted analysis where no prior selection of pathways is performed, which is a hypothesis-driven approach. Both have advantages and disadvantages, and the approach taken ultimately depends on the study in question. In this introductory chapter, we outline the major challenges involved with such an approach. Subsequent chapters provide detailed discussions of the crucial points.
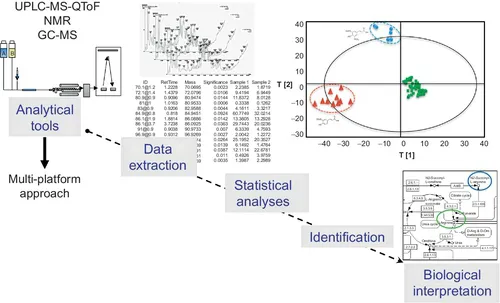
Figure 1.2 Workflow of a metabolomic analysis.
1.2 The experimental design
So far, metabolomic studies have mainly used animal nutritional interventions and human clinical trials. More recently, cohorts with 7–12 year follow-up have largely been utilised for discovering predictive biomarkers for the evolution of a health state towards a disease state. These studies are described in Chapter 6. As previously reported, metabolomics was first developed and applied to toxicology and pharmacology, which usually induce major changes in the blood or urine metabolome after the administration of a drug (Nicholson et al., 2002; Pujos-Guillot et al., 2012). This is not the case when applying metabolomics to nutrition research because dietary interventions most often generate subtle changes in metabolic profiles, which may sometimes be difficult to detect (Solanky et al., 2003). Considering interindividual response to diet (Walsh et al., 2006), the effects of many factors, such as age, sex, and various genetic and environmental stimuli on the metabolic profiles of the biofluids (Johnson and Gonzalez, 2011), researchers must take great care when designing the experimental protocol of a metabolomics approach, especially when using humans, as subtle variations between subjects may induce large variations in metabolite concentrations in the plasma and/or urine. Therefore, the protocol should be designed to reduce confounders as much as possible, allowing retrieval of optimised information from the analytical data (Smilowitz et al., 2013).
For metabolomic studies of predictive biomarkers, the use of cohorts and epidemiological approaches benefit from very comprehensive data sets that have been acquired during the follow-up years. These data include clinical measurements (blood and urine biochemistry, anthropometric and body-composition measurements), number of events (cardiovascular diseases, type 2 diabetes, etc.), food frequency questionnaires, socioeconomic data, and blood and urine samples for further metabolomic analyses. One of the major challenges of this type of study is to correctly evaluate the dietary intake of the individuals. At the moment, 24-h recalls, weighed food diaries, and food frequency questionnaires are being used, but the issues surrounding their use are well documented (Fave et al., 2009). Consequently, researchers are trying to use metabolomics to identify the food metabolome (Llorach et al., 2012; Pujos-Guillot et al., 2013), in order to evaluate dietary exposure based on markers of specific food intake (Fave et al., 2009). Chapter 9 discusses this important topic.
1.3 The analytical platform
Human biofluids are a complex mixture of thousands of metabolites that include endogenous and exogenous molecules, as reported for se...
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- List of contributors
- Woodhead Publishing Series in Food Science, Technology and Nutrition
- Preface
- Part One: Principles
- Part Two: Applications in nutrition research
- Index