
- 334 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Clinical Applications for Next-Generation Sequencing
About this book
Clinical Applications for Next Generation Sequencing provides readers with an outstanding postgraduate resource to learn about the translational use of NGS in clinical environments.Rooted in both medical genetics and clinical medicine, the book fills the gap between state-of-the-art technology and evidence-based practice, providing an educational opportunity for users to advance patient care by transferring NGS to the needs of real-world patients.The book builds an interface between genetic laboratory staff and clinical health workers to not only improve communication, but also strengthen cooperation. Users will find valuable tactics they can use to build a systematic framework for understanding the role of NGS testing in both common and rare diseases and conditions, from prenatal care, like chromosomal abnormalities, up to advanced age problems like dementia.- Fills the gap between state-of-the-art technology and evidence-based practice- Provides an educational opportunity which advances patient care through the transfer of NGS to real-world patient assessment- Promotes a practical tool that clinicians can apply directly to patient care- Includes a systematic framework for understanding the role of NGS testing in many common and rare diseases- Presents evidence regarding the important role of NGS in current diagnostic strategies
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
Next Generation Sequencing—General Information about the Technology, Possibilities, and Limitations
Abstract
Keywords
Enrichment; Illumina; Next generation sequencing; NGS platforms; PacBio; PGM; Proton Ion; Whole exome sequencing; Whole genome sequencingNGS Versus Traditional (Sanger Sequencing)
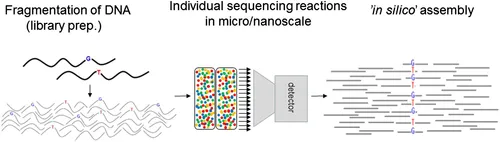
Coverage
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- List of Contributors
- Chapter 1. Next Generation Sequencing—General Information about the Technology, Possibilities, and Limitations
- Chapter 2. Basic Bioinformatic Analyses of NGS Data
- Chapter 3. Analysis of Structural Chromosome Variants by Next Generation Sequencing Methods
- Chapter 4. Next Generation Sequencing in Oncology
- Chapter 5. Next Generation Sequencing in Hematological Disorders
- Chapter 6. Next Generation Sequencing in Neurology and Psychiatry
- Chapter 7. Next Generation Sequencing in Dysmorphology
- Chapter 8. Next Generation Sequencing in Vision and Hearing Impairment
- Chapter 9. Next Generation Sequencing as a Tool for Noninvasive Prenatal Tests
- Chapter 10. Clinical Applications for Next Generation Sequencing in Cardiology
- Chapter 11. Next Generation Sequencing in Pharmacogenomics
- Chapter 12. The Role of Next Generation Sequencing in Genetic Counseling
- Chapter 13. Next Generation Sequencing in Undiagnosed Diseases
- Chapter 14. Organizational and Financing Challenges
- Chapter 15. Future Directions
- Chapter 16. Ethical and Psychosocial Issues in Whole-Genome Sequencing for Newborns
- Chapter 17. Next Generation Sequencing—Ethical and Social Issues
- Index
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app