![]()
1
MHC and T cell responses
Abstract:
The major histocompatibility complex (MHC) molecules of classes I and II, coded by the human leukocyte antigen (HLA) genes on chromosome 6, are cell surface glycoproteins that play a major role in T cell-mediated immune recognition. Class I molecules are found on the surface of virtually all nucleated cells in humans, except neurons, and are involved in detection of viral infections in cells. Class II molecules are limited to specific antigen presenting cells. In both cases, T cell antigen recognition is triggered by the MHC presenting a short antigenic peptide fragment which binds to a specific T cell receptor (TCR) on the surface of T cells. In this chapter, we survey the nature of the MHC molecules and their roles in cell-mediated immune responses. The genetic organizations of the MHC class I and II molecules, their structures and functions are described.
Key words
major histocompatibility complex
human leukocyte antigen
T cell
T cell receptor
antigen presenting cell
cell-mediated immunity
1.1 Genetic organization of the MHC
The major histocompatibility complex (MHC) molecules are cell surface glycoproteins that play a central role in T cell-mediated immune recognition. The MHC genes in humans, termed human leukocyte antigen (HLA), are found on chromosome 6. Today, HLA is organized into three major genetic regions or loci designated class I, II and III. Class III genes primarily encode components of the serum complement system. Class I and class II loci, on the other hand, encode a number of highly polymorphic cell-surface proteins responsible for antigen presentation. The HLA class I locus is subdivided into HLA-A, -B, and -C subregions, each encoding class I α chain genes. The class II HLA locus, HLA-D, is subdivided into at least six subregions, namely HLA-DR, -DQ, -DP, -DX, -DO, and -DZ. The class I and class II genes are highly polymorphic genes in the human genome; for some of these genes, over 200 allelic variants have been identified. HLA specificities are identified by an identifier for locus and a number (e.g. A1, DR4, and DQ5) and the haplotypes are identified by individual specificities. Specificities that are defined by genomic analysis are names beginning with an identifier for the locus followed by a four-digit code (e.g. A*0101, Cw*0401, and DRB1*0503). Despite considerable MHC polymorphism, a single individual expresses a finite number of MHC alleles and is heterozygous for each MHC gene in humans. The work discussed here focuses on MHC molecules that are responsible for antigen presentation. Therefore, the use of MHC for the rest of the text is restricted to only the class I and class II genes.
1.2 MHC structure
All MHC molecules share certain structural characteristics that are critical for their role in peptide display and recognition by T cells. T cell recognition of antigen is said to be MHC restricted, as T cell receptors (TCRs) will only bind to fragments of antigen that are associated with products of the MHC. Each MHC molecule contains an extracellular peptide-binding cleft which is composed of paired α-helices resting on a floor consisting of an eight-stranded anti-parallel β-sheet. This portion of the MHC molecule binds antigenic peptides for display to T cells, and the TCRs possess a complementary shape that will interact with the displayed peptide and with the helices of the MHC molecules. The amino acid residues located in and around this cleft are highly polymorphic and they are responsible for the different peptide binding specificities among different MHC alleles. A non-polymorphic determinant on the MHC molecules acts as the binding site for the T cell co-receptor molecules CD4 and CD 8. CD4 and CD 8 are expressed on distinct subpopulations of mature T cells and, together with the antigen receptors, participate in the recognition of antigen. CD8 binds selectively to class I MHC molecules, and CD4 binds to class II MHC molecules. Hence, CD8+ T cells recognize only peptides displayed by class I molecules, and CD4+ T cells recognize only peptides presented by class II molecules. Most CD8+ T cells function as cytotoxic T cells and CD4+ cells are helper cells.
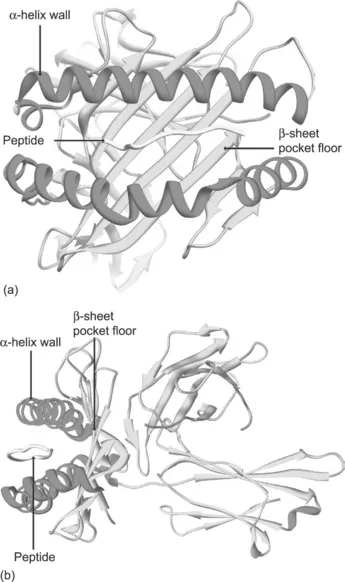
MHC class I molecules are ternary complexes composed of a heavy glycosylated transmembrane protein non-covalently linked to a smaller polypeptide β2-microglobulin (β2m). The complete molecule has four globular domains: three formed by the heavy chain (α1, α2, α3) and one by β2m, as shown in Figure 1.1. Both the α1 and α2 domains adopt a similar structure: starting from the N-terminus, each region of the chain forms four anti-parallel β-strands followed by a helical region across the β-strands on one side of the β-sheet. The two domains associate in such a way that their β-sheets are hydrogen-bonded to each other, forming a platform of a continuous eight-stranded anti-parallel β-sheet. The β-sheet is relatively flat with a small propeller twist. The sides of this cleft are formed by two α-helices, one from α1 and one from α2. It is within this cleft that antigen fragments are held and presented to T cells. The α3 domain consists of a transmembrane segment and a short cytoplasmic tail that anchors the molecule in the membrane.
Figure 1.1 Schematic of MHC class I structure. (a) Front view of a class I molecule in complex with antigenic peptide based on X-ray crystallography structure. (b) Side view of the same molecule clearly showing the anatomy of the peptide-binding cleft formed by α-helices sitting on a platform of a β-sheet
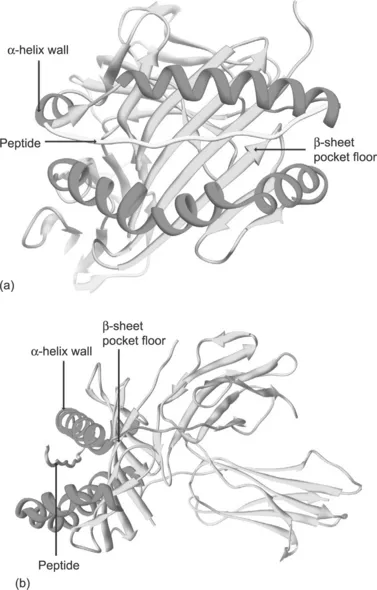
MHC class II molecules are also transmembrane glycoproteins, consisting of two polypeptide chains (α, β) held together by non-covalent interactions. Similarly to class I MHC, the complete class II MHC molecule has four globular domains, two on each chain (α1, α2, β1, β2). The α1 and α1 domains mimic the class I α1 and α2 domains in forming a peptide-binding groove bounded by two a-helices and a β-sheet floor (Figure 1.2).
Figure 1.2 Schematic of MHC class II structure. (a) Front view of a MHC class II molecule in complex with antigenic peptide extending out of the binding cleft based on X-ray crystallographic structure. (b) Side view of the same molecule showing the anatomy of the peptide-binding cleft formed by α-helices sitting on a platform of β-sheet
1.3 MHC function
The class I MHC-restricted antigen processing and presentation pathway provides a sophisticated surveillance mechanism aimed at detecting viral infections in cells. MHC class I molecules are synthesized in the endoplasmic reticulum (ER) and are present on the surface of virtually all nucleated cells, except neurons, in humans. Their function is to bind peptides derived from endogenous antigens within the cell, transport them to the cell surface, and present the bound peptide ligands to cytotoxic T cells through the TCR and CD8. Most class I peptide ligands are derived from proteins that are degraded by proteasomes. The proteasome has broad substrate specificity and can generate a wide variety of peptides from cytosolic proteins. Exposure of cells to interferon (IFN)-γ induces the synthesis of three proteolytic proteasome subunits - low molecular weight proteins (LMP)-2 and LMP-7 and multicatalytic endopeptidase complex (MECL)-1 – which are incorporated into an alternative form of proteasome, called an immunoproteasome, displacing the constitutive subunits β1, β2, and β5, respectively. At the present time, it remains unclear how the products of such endopeptidase activity are related to the final MHC class I ligands. One possibility is that the proteasomes directly produce peptides of appropriate size. Alternatively, the proteasomes may generate longer peptides that require further processing. It is also possible that two short non-continuous peptide fragments can be fused together to create the final class I ligand via post-translational protein splicing. In any case, the majority of these peptides are transported from the cytosol into the ER by the transporter associated with antigen processing (TAP). TAP consists of two structurally related subunits, which interact to form a functional peptide-transporting complex. Before peptide translocation by TAP, peptides bind to the membrane- proximal, cytosolic surface of TAP1/TAP2 complexes. Hydrolysis of ATP results in peptide translocation into the ER lumen. Within the ER lumen, precursor peptides may be further trimmed by an ER-resident aminopeptidase ERAAP (ER aminopeptidase associated with antigen processing) before loading onto MHC class I molecules. The class I peptide/MHC complexes eventually exit the ER by association with B cell-associated protein Bap31.
Similarly to MHC class I molecules, MHC class II molecules are also synthesized in the ER. In addition to the polypeptide α and β chains, an invariant chain (Ii) is also produced within the ER, which associates with MHC class II molecules before they reach the cell surface. Unlike MHC class I expression, which encompasses most cells, class II MHC expression is limited to specific antigen presenting cells (APCs) such as dendritic cells, endothelial cells, monocytes and B cells. They present exogenous peptide antigens to helper T cells through the TCR and CD4. Exogenous foreign antigen is processed through the MHC class II pathway. Antigen is internalized and degraded enzymatically in endosomes and lysosomes into peptide fragments. MHC class II molecules remain competent for peptide loading by binding fragments of Ii in the ER. These fragments remain...