
- 488 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Proteomic and Metabolomic Approaches to Biomarker Discovery
About this book
Proteomic and Metabolomic Approaches to Biomarker Discovery demonstrates how to leverage biomarkers to improve accuracy and reduce errors in research. Disease biomarker discovery is one of the most vibrant and important areas of research today, as the identification of reliable biomarkers has an enormous impact on disease diagnosis, selection of treatment regimens, and therapeutic monitoring. Various techniques are used in the biomarker discovery process, including techniques used in proteomics, the study of the proteins that make up an organism, and metabolomics, the study of chemical fingerprints created from cellular processes.Proteomic and Metabolomic Approaches to Biomarker Discovery is the only publication that covers techniques from both proteomics and metabolomics and includes all steps involved in biomarker discovery, from study design to study execution. The book describes methods, and presents a standard operating procedure for sample selection, preparation, and storage, as well as data analysis and modeling. This new standard effectively eliminates the differing methodologies used in studies and creates a unified approach. Readers will learn the advantages and disadvantages of the various techniques discussed, as well as potential difficulties inherent to all steps in the biomarker discovery process.A vital resource for biochemists, biologists, analytical chemists, bioanalytical chemists, clinical and medical technicians, researchers in pharmaceuticals, and graduate students, Proteomic and Metabolomic Approaches to Biomarker Discovery provides the information needed to reduce clinical error in the execution of research.- Describes the use of biomarkers to reduce clinical errors in research- Includes techniques from a range of biomarker discoveries- Covers all steps involved in biomarker discovery, from study design to study execution
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
Biomarker Discovery
Study Design and Execution
Abbreviations
Acknowledgments
Introduction
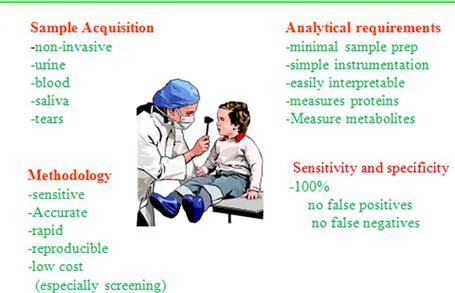
Definitions
Biomarker
Sensitivity
Specificity
Positive Predictive Value (PPV)
Negative Predictive Value (NPV)
Proteomics
Profiling
Metabolomics
The Current State of Biomarker Discovery
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Preface
- List of Contributors
- Chapter 1. Biomarker Discovery: Study Design and Execution
- Chapter 2. Proteomic and Mass Spectrometry Technologies for Biomarker Discovery
- Chapter 3. Tissue Sample Preparation for Proteomic Analysis
- Chapter 4. Sample Preparation in Global Metabolomics of Biological Fluids and Tissues
- Chapter 5. Serum and Plasma Collection: Preanalytical Variables and Standard Operating Procedures in Biomarker Research
- Chapter 6. Current NMR Strategies for Biomarker Discovery
- Chapter 7. Using Data-Independent Mass Spectrometry to Extend Detectable Dynamic Range without Prior Fractionation
- Chapter 8. Gas Chromatography/Mass Spectrometry-Based Metabonomics
- Chapter 9. Liquid Chromatographic Methods Combined with Mass Spectrometry in Metabolomics
- Chapter 10. Capillary Electrophoresis–Mass Spectrometry for Proteomic and Metabolic Analysis
- Chapter 11. Current Gel Electrophoresis Approaches to Low-Abundance Protein Marker Discovery
- Chapter 12. Two-Dimensional Difference in Gel Electrophoresis for Biomarker Discovery
- Chapter 13. Affinity Targeting Schemes for Biomarker Research
- Chapter 14. Asp-Selective Microwave-Supported Acid Proteolysis
- Chapter 15. Sample Depletion, Fractionation, and Enrichment for Biomarker Discovery
- Chapter 16. Protein and Metabolite Identification
- Chapter 17. Quantitative Proteomics in Development of Disease Protein Biomarkers
- Chapter 18. Mass Spectrometry and NMR Spectroscopy–Based Quantitative Metabolomics
- Chapter 19. Multivariate Analysis for Metabolomics and Proteomics Data
- Chapter 20. Top-Down Mass Spectrometry for Protein Molecular Diagnostics and Biomarker Discovery
- Chapter 21. A Role for Protein–Protein Interaction Networks in the Identification and Characterization of Potential Biomarkers
- Chapter 22. Reverse Phase Protein Microarray Technology: Advances into the Clinical Research Arena
- Chapter 23. Autoantibodies and Biomarker Discovery
- Chapter 24. MicroRNAs and Biomarker Discovery
- Chapter 25. Imaging Mass Spectrometry of Intact Biomolecules in Tissue Sections
- Chapter 26. Mass Spectrometry–Based Approach for Protein Biomarker Verification
- Chapter 27. Mass Spectrometry Metabolomic Data Handling for Biomarker Discovery
- Chapter 28. Analytical Methods and Biomarker Validation
- Index
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app