
- 482 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Calmodulin and Signal Transduction
About this book
Calmodulin and Signal Transduction focuses on emerging themes in the molecular mechanisms of calcium signal transduction through calmodulin-regulated pathways. It provides the reader with selected examples and experimental precedents that underlie current models of cell regulation through calmodulin-regulated pathways and their linkage with other regulatory pathways.
- Molecular mechanisms of calcium signal transduction through calmodulin-regulated enzymes
- Selected case studies and precedents related to molecular mechanisms
- Protein-protein recognition between calmodulin and the enzymes it regulates
- Cross-talk and interdigitation with other signal transduction pathways
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Calmodulin and Signal Transduction by Linda J. Van Eldik,D. Martin Watterson in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Physiology. We have over one million books available in our catalogue for you to explore.
Information
1
Calmodulin and Calcium Signal Transduction: An Introduction
LINDA J. VAN ELDIK*† and D. MARTIN WATTERSON*‡
*Northwestern Drug Discovery Program
†Departments of Cell and Molecular Biology
‡Molecular Pharmacology and Biological Chemistry, Northwestern University Medical School, Chicago, Illinois 60611
I. Background
II. Phylogenetic Conservation of Calmodulin
III. Molecular Mechanisms of Calmodulin-Mediated Ca2+ Signal Transduction
References
I. BACKGROUND
Calcium has diverse and broad roles in nature. This book is concerned with only a small part of the world of calcium—its functioning as a critical component of a dynamic system in eukaryotic cells that uses the protein calmodulin (CaM) as a calcium sensor and transducer. Calmodulin has the broadest phylogenetic and ontogenetic distribution in its class of eukaryotic proteins and is the most extensively studied. Calmodulin, therefore, has been called a prototype for eukaryotic cell Ca2+ signal transduction and homeostasis. The mechanism by which CaM is able to transduce Ca2+ signals into biological responses is through its modulation of the structure and function of other proteins that usually lack the ability to bind Ca2+ with the affinity and kinetics required for the biological response (see chapter by Nelson and Chazin). Most of these CaM-modulated proteins are enzymes (see chapters by Lukas et al.; Perrino and Soderling; Sonnenburg et al.; Hu and Van Eldik; Brandt and Vanaman). This provides the potential for CaM to amplify, through the regulation of enzyme-based catalysis, a stoichiometric bioinorganic equilibrium into a series of reiterative chemical reactions. Because CaM is the calcium sensor for multiple enzymes, there is amplification of diverse chemical reactions in response to a given Ca2+ signal. In addition to enzyme-based catalysis, the cytoskeleton is an area of CaM regulation that is receiving increasing attention and provides a different perspective on how CaM can be involved in cellular homeostasis (see chapter by Bonafé and Sellers). Because many cytoskeletal proteins serve as both structural proteins and regulatory systems, they offer another level, or mode, of regulation. If CaM is involved in the regulation of nodes in an extended array of biopolymers, then there is the opportunity to transduce a signal through a physical transducer system, rather than through an enzymatic amplification system. All in all, CaM itself appears to be an integrator through its presence as a calcium sensor in a diverse array of regulatory pathways.
Calmodulin activity was discovered in the early 1970s when the late W. Y. (George) Cheung and S. Kakiuchi independently described (Cheung, 1970; Kakiuchi and Yamazaki, 1970) an activating factor for cyclic nucleotide phosphodiesterase (PDE). A few years later Wang and co-workers (Teo and Wang, 1973) showed that the PDE-activating factor required calcium for activity. An early example of diverse fields of research converging at the subject of CaM research was with the first reports of the purification of CaM to chemical homogeneity (Stevens et al., 1976; Watterson et al., 1976). Stevens et al. (1976) isolated the protein as part of attempts to purify and characterize the PDE activator protein and noted that it had some similarities to troponin C from vertebrate skeletal muscle. Watterson et al. (1976) purified the CaM to homogeneity in their search for nonmuscle proteins that had physical, chemical, and calcium-binding properties similar to those of skeletal muscle troponin C. The search was not to find a protein that would function as a troponin C, but to find a new class of proteins that would have the general calcium-binding properties required to serve as a calcium signal transducer. Troponin C was the best characterized calcium signal transducer protein at the time and served as the standard of comparison. Since that time, CaM has become the standard of comparison, with troponin Cs being viewed as tissue-specific CaM-like proteins.
With the availability of a homogeneous protein, there was further convergence of diverse fields as several investigators proceeded to show that the calcium-binding subunit, or calcium-activating factor, of other enzymes was indistinguishable from this new standard of comparison, which had not yet been named CaM. By the late 1970s, it was apparent that there were several enzymes that were probable physiological targets for CaM. In addition to PDE (see chapter by Sonnenburg et al.), these included adenylyl cyclase (see chapter by Sonnenburg et al.), the ATPase pump (see chapter by Brant and Vanaman), myosin light chain kinase (see chapter by Lukas et al.), and phosphorylase kinase (see chapter by Lukas et al.). The name calmodulin, derived from calcium modulator protein, was coined by Cheung in 1978 (Cheung et al., 1978) to reflect the multifunctional nature of CaM as a calcium regulatory protein. It was at this time that the dogma of CaM being present in a great molar excess over its targets became established. This was based on the observation that the concentration of CaM in tissues such as vertebrate brain far exceeded the estimated concentrations of the limited number of enzymes known at the time. However, the number of CaM targets proteins was soon to be expanded.
II. PHYLOGENETIC CONSERVATION OF CALMODULIN
The first complete amino acid sequence of CaM (Watterson et al., 1980) was reported in 1980. The 1980s witnessed the expansion of the field to phylogenetics with the purification and characterization of CaMs from diverse species. For example, the first amino acid sequences of CaMs from higher plants (Lukas et al., 1984), a slime mold (Marshak et al., 1984), a unicellular alga (Schleicher et al., 1984; Zimmer et al., 1988), a ciliated protozoan (Schaefer et al., 1987), the fruit fly Drosophila (Smith et al., 1987; Yamanaka et al., 1987), and yeast (Davis et al., 1986) were reported. It was the phylogenetic conservation of residues among diverse CaMs with retention of in vitro activity that provided the basis of the first site-directed mutagenesis and chimeric CaM analyses (Roberts et al., 1985; Craig et al., 1987; Putkey et al., 1988). As discussed in the following chapters in this volume, the conclusions from these results were consistent with the structures of CaM:peptide complexes reported several years later, and the two types of approaches are proving to be complementary in the dissection of the detailed relationships among structures and functions (see chapter by Lukas et al.).
Based on the large number of biochemical characterizations of CaMs from multiple species and analysis of the amino acid variability allowed in the CaM sequence for isofunctionality, the current state of knowledge about CaM sequences can be summarized in Fig. 1. A generic CaM sequence and the vertebrate CaM sequence are shown. Calmodulin is one of the most highly conserved eukaryotic proteins known. The positions of selected conserved residues are landmarked in the three-dimensional structure of CaM shown in Fig. 2. In contrast to the high conservation of the amino acid sequence throughout species, the CaM gene varies among eukaryotes in both the number of genes and the structural organization. Calmodulin can be encoded by a single gene in an organism or by multiple genes, the number and position of introns differ among CaM genes, and the length of 5′ and 3′ flanking regions varies. Although the number and structural organization of CaM genes vary among species, these genes encode identical amino acid sequences in most cases. The highly conserved primary structure of CaM may reflect the necessity for a highly conserved structure for highly conserved functions or may reflect the importance of CaM as a multifunctional transducer of calcium signals through target protein regulation and the need to conserve a diversity of recognition interfaces in a single, relatively small protein.
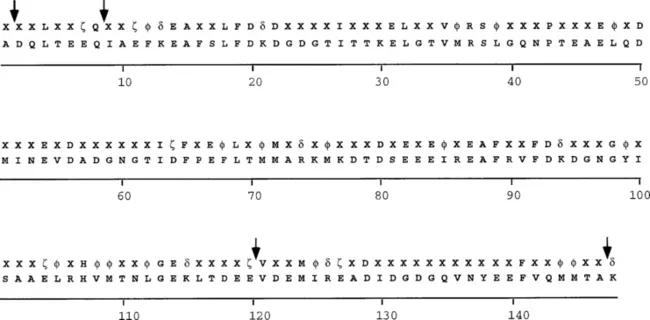
Fig. 1 Consensus sequence in naturally occurring CaMs. A consensus sequence (upper rows) and a vertebrate CaM sequence (lower rows) are shown. For the vertebrate CaM sequence, standard single-letter amino acid codes are used. To evaluate the CaM consensus sequence, sequences of naturally occurring CaMs obtained from the Swiss-Prot or GenBank databases were aligned: φ, Hydrophobic amino acids (I,L,V,M,F,Y); ζ, negatively charged amino acids (D,E); δ, positively charged amino acids (K,R); and X, any amino acid. Positions where amino acid insertions are found in some CaMs are indicated by arrows. Amino acid numbering is that from vertebrate CaM.
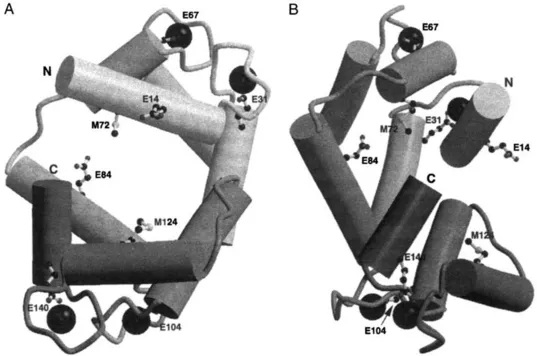
Fig. 2 Phylogenetically conserved amino acid residues in the three-dimensional structure of CaM. These ribbon drawings are based on coordinates in the Brookhaven Protein Data Bank (PDB) entry 1CDL and represent one of the multiple conformations that CaM can assume. Two different views of CaM are shown, with side chains of selected amino acids that are phylogenetically conserved and noted in Fig. 1. Ca2+ ions are solid spheres. The amino (N) and carboxy (C) terminal ends of CaM are indicated.
III. MOLECULAR MECHANISMS OF CALMODULIN-MEDIATED Ca2+ SIGNAL TRANSDUCTION
Concurrent with the study of CaM biochemistry and genetics, progress was being made in the probing of the initial steps in the molecular mechanism of CaM action. The development of CaM-Sepharose chromatography (Watterson and Vanaman, 1976) in the 1970s provided a facile and gentle step late in purifications of CaM-regulated proteins, resulting in an increased number of purified CaM-binding proteins and the analysis of the next step in the mechanism. Although efficient use of the calcium-dependent chromatography step required the prior depletion of the endogenous CaM, or CaM-like protein, by differential centrifugation or ion-exchange chromatography, there was a tendency to equate the biochemical properties with the physiological state of the calcium signal transduction complex. From the 1980s and well into the 1990s, new CaM-binding proteins were discovered as described in the following chapters in this volume. The focus on characterizing the components of Ca2+ signal transduction pathways continued to bring investigators in diverse fields together as they found a CaM-regulated protein as a component of their pathway of interest. The concept of CaM being the calcium-binding subunit of a diversity of CaM-regulated enzymes and a subunit of abundant regulatory proteins, such as the myosins, in the same tissue or cell is now generally accepted. This requires that the question of molar stoichiometry be revisited, especially if the field is to move to the next level of research detail in which pathways are modeled as a basis for forecasting the response of cells to a given stimulus. In this regard, the field of CaM research is poised to take advantage of the emerging discipline of bioinformatics and its application to drug discovery and the analysis of signal transduction homeostasis.
Based on the increasing database of knowledge about CaM and its targets, there are a number of general principles and emerging themes concerning the molecular mechanisms of action of CaM as a transducer o...
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Contributors
- Preface
- Chapter 1: Calmodulin and Calcium Signal Transduction: An Introduction
- Chapter 2: Calmodulin as a Calcium Sensor
- Chapter 3: Calmodulin-Regulated Protein Kinases
- Chapter 4: Biochemistry and Pharmacology of Calmodulin-Regulated Phosphatase Calcineurin
- Chapter 5: Cyclic Nucleotide Regulation by Calmodulin
- Chapter 6: Regulation of Nitric Oxide Synthase by Calmodulin
- Chapter 7: Calmodulin-Binding Proteins of the Cytoskeleton
- Chapter 8: Calmodulin and Ion Flux Regulation
- Index