
- 612 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
From Gene to Protein: Translation into Biotechnology
About this book
From Gene to Protein: Translation into Biotechnology is the 15th volume in the continuing series under the title ""Miami Winter Symposia"". The theme of the symposium is the translation of the basic research findings into the practical application of biotechnology. This book summarizes methodology and its applications that lie behind the practical innovations. The book starts with reviews of techniques of eukaryotic cell culture, hybridoma technology and uses, and the in vitro synthesis of DNA and its use in the generation of protein analogs. Considerable space is devoted to development of monoclonal antibodies that promises to be the dominating tool of medical technology, both for diagnosis and therapy. Cloning into eukaryotic cells and methods of increasing the levels of gene expression are included. These topics reflect areas of intensive research that have important commercial and clinical value. Core chapters describe biological activities of cloned gene products, including reports on trials with human subjects of interferon, human insulin, and growth hormone. A panel session on horizons in biotechnology is also provided, looking forward to the directions of future research and its applications. Biotechnologists, cell biologists, scientists, researchers, teachers, and students will greatly benefit from this book.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access From Gene to Protein: Translation into Biotechnology by Fazal Ahmad in PDF and/or ePUB format, as well as other popular books in Physical Sciences & Industrial & Technical Chemistry. We have over one million books available in our catalogue for you to explore.
Information
Biological Activities of Cloned Gene Products
REGULATION OF HISTONE GENE EXPRESSION IN HUMAN CELLS
G.S. Stein , J.L. Stein*, L. Baumbach , A. Leza*, A. Lichtler , F. Marashi , M. Plumb , R. Rickles , F. Sierra and T. Van Dyke , Department of Biochemistry and Molecular Biology, University of Florida, Gainesville, Florida; *Department of Immunology and Medical Microbiology, University of Florida, Gainesville, Florida
Publisher Summary
Histone genes represent a moderately repeated set of genes in human cells. To study the organization and regulation of human histone genes, we have characterized a series of recombinant lambda Charon 4A phage containing genomic human histone sequences (designated λHHG). Our analyses indicate that human histone genes are clustered but are not organized in a simple tandem repeat pattern as is observed for several lower eukaryotes. For example, several of the human genomic fragments we have isolated contain two each of segments coding for H3 and H4 histones. Of particular interest with respect to organization and expression of the human histone genes is the presence of at least seven different H4 histone mRNAs associated with polysomes of S phase HeLa cells. At least three of the HeLa H4 histone mRNAs are products of distinct genes and two H4 histone genes in the same genomic fragment code for different H4 mRNAs.We have used our cloned human histone genes to examine the regulation of histone gene expression in human cells. Though it is generally agreed that histone protein synthesis in HeLa cells is restricted to the S phase of the cell cycle, and therefore parallels DNA replication, both transcriptional and post-transcriptional levels of control have been postulated. By probing electrophoretically fractionated, filter-immobilized RNAs with several cloned genomic human histone sequences representing different histone gene clusters, we have assessed the steady state levels of histone RNAs in the nucleus and cytoplasm of G1 and S phase HeLa S3 cells. The representation of histone mRNA sequences in G1 compared with S phase cells was less than 1% in the cytoplasm and approximately 1% in the nucleus. These data are consistent with control occurring primarily at the transcriptional level, but we cannot dismiss the possibility that regulation of histone gene expression is, to some extent and/or under some biological circumstances, mediated post-transcriptionally. If histone gene transcription does occur in G1, the RNAs must either be rapidly degraded or be transcribed to a limited extent compared with S phase.An unexpected result was obtained when a northern blot of cytoplasmic RNA from G1 and S phase cells was hybridized with λHHG41 DNA (containing H3 and H4 genomic human histone sequences). This clone hybridizes with histone mRNAs present in S phase cytoplasmic RNA, but also hybridizes with a G1 cytoplasmic RNA approximately 330 nucleotides in length. This RNA, present in the cytoplasm of HeLa cells predominantly in the G1 phase of the cell cycle, is not similar in size or nucleotide sequence to known histone RNAs.The possibility of prokaryotic-like organization and regulation of human histone genes is discussed.
I. INTRODUCTION
Histone genes are represented as a moderately repeated set of genomic DNA sequences in human cells, and compelling evidence points towards a major role for the gene products, the histone proteins, in packaging newly replicated DNA and in modifications of genome structure associated with transcription. In this chapter we will summarize the progress our laboratory has made during the past several years towards addressing the structural and functional properties of human histone genes: First, the structure and organization of human histone genes will be discussed, primarily because of the functional interrelationships between structure, organization and regulation but also because of the utilization of specific regions of genomic histone sequences as probes for analysis of human histone gene organization and regulation of expression. Second, approaches to assessing levels of control of histone gene expression will be considered, and here, variations in levels of control under different biological circumstances and the possibility of prokaryotic-type organization and regulation of human histone gene will be evaluated.
II. STRUCTURE AND ORGANIZATION OF HUMAN HISTONE GENES
We have isolated a series of twelve genomic clones containing human histone coding sequences, their flanking sequences and noncoding sequences from a λ Charon 4A human gene library constructed by Tom Maniatis and collaborators (1). These clones were analyzed by hybridization with heterologous probes as well as by hybrid selection-translation (Figure 1A), nucleotide sequencing (Figure 1B) and restriction enzyme mapping (39). While the moderately repeated human histone genes are clustered, they are not arranged in the form of a tandem repeat such as has been observed in sea urchin and Drosophila. Rather, human histone genes exhibit at least three types of arrangements with respect to restriction sites and the order of coding sequences; each of these arrangements is clearly distinguishable from the others. The organization of human histone genes is further complicated by the association of Alu family DNA sequences with some but not all of the histone coding regions (40). Additionally, sequences coding for non-histone RNA species, some of which are expressed throughout the cell cycle and others which are expressed during specific periods of the cell cycle, are interspersed among the human histone genes. In situ hybridization studies (2,3) and restriction analysis data from human-rodent hybrids (4) suggest that in humans the histone genes may be clustered on the distal end of the long arm of chromosome 7 in the G-negative band q34. Partial restriction maps of each of the three types of arrangements of histone genes, representing information from seven individual genomic clones, are shown in Figures 1C and 1D.
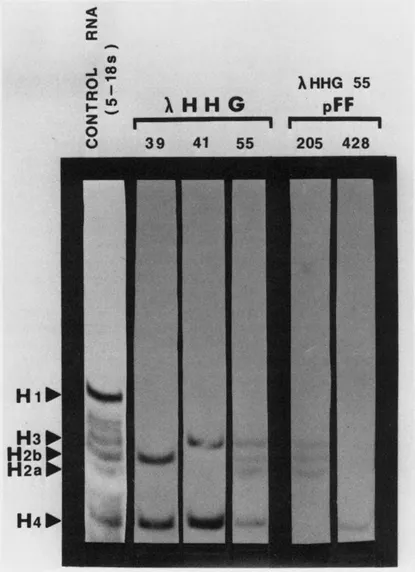
Figure 1A Hybrid selection-in vitro translation analysis of DNAs from λHHG recombinant phage-containing human histone sequences and of recombinant pBR322 plasmids containing specific regions of the genomic human histone sequences subcloned from λHHG phage. Phage or plasmid DNA was immobilized on nitrocellulose filters and hybridized with HeLa S phase polysomal RNA containing H2A, H2B, H3, H4 and H1 histone mRNAs. Hybridized RNAs were eluted, translated in a wheat germ cell-free protein synthesizing system, and the in vitro translated polypeptides were fractionated electrophoretically in acetic acid-urea polyacrylamide gels. Analysis was by autoradiography. The left lane shows in vitro translation products (all five histone polypeptides) translated from the RNA preparation used for hybrid selection.

Figure 1B Partial nucleotide sequence analysis of the human H3 histone gene in the 3.1 Kb Eco RI fragment of λHHG17. Shown for comparison are the nucleotide sequences of several other H3 genes. The asterisk (*) indicates a codon which would result in an amino acid substitution.
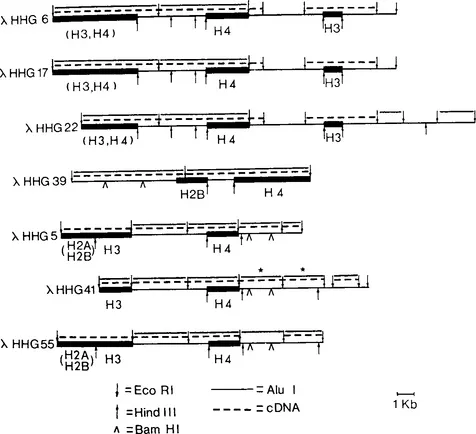
Figure 1C Partial restriction maps of seven recombinant λ phages containing genomic human histone sequences. Also designated are regions of the phage which contain Alu family DNA sequences and nonhistone coding regions which hybridized with cDNA complementary to S phase polysomal RNAs.
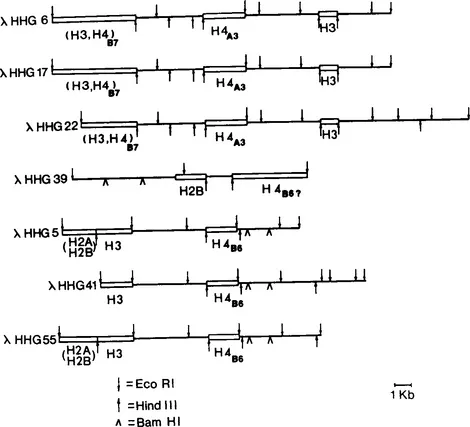
Figure 1D Partial restriction maps of the λHHG phage in which the variant H4 histone mRNA coding sequences are designated.
Of particular interest with respect to the structure, organization and expression of human histone genes is an observation we made severa...
Table of contents
- Cover image
- Title page
- Table of Contents
- MIAMI WINTER SYMPOSIA—VOLUME 19
- Copyright
- SPEAKERS AND DISCUSSANTS
- PREFACE
- The Thirteenth Lynen Lecture
- Introduction
- Techniques of Eukaryotic Cell Culture
- Monoclonal Antibodies—Production and Uses
- In Vitro Synthesis of DNA and the Generation of Protein Analogs
- Cloning into Eukaryotic Cells
- Increasing Levels of Gene Expression
- Biological Activities of Cloned Gene Products
- Horizons in Biotechnology
- Free Communications
- INDEX OF AUTHORS
- SUBJECT INDEX