
- 826 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Principles of Bacterial Pathogenesis
About this book
Principles of Bacterial Pathogenesis presents a molecular perspective on a select group of bacterial pathogens by having the leaders of the field present their perspective in a clear and authoritative manner. Each chapter contains a comprehensive review devoted to a single pathogen. Several chapters include work from authors outside the pathogenesis field, providing general perspectives on the evolution, regulation, and secretion of virulence and determinants.
- Explains the basic principles of bacterial pathogenesis
- Covers diverse aspects integrating regulation, cellular microbiology and evolution of microbial disease of humans
- Discusses current strategies for the identification of virulence determinants and the methods used by microbes to deliver virulence factors
- Presents authoritative treatises of the major disease microorganisms
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Principles of Bacterial Pathogenesis by Eduardo A. Groisman in PDF and/or ePUB format, as well as other popular books in Medicine & Infectious Diseases. We have over one million books available in our catalogue for you to explore.
Information
Topic
MedicineSubtopic
Infectious DiseasesCHAPTER 1
Evolution of Bacterial Pathogens
HOWARD OCHMAN
I. Introduction
II. The Genetic Basis of Virulence
III. Identification of Sequences Involved in Bacterial Pathogenesis
IV. Recovery of Genes Contributing to Virulence
V. The Population Genetics of Pathogens
VI. Studying Bacterial Population Genetics
A. Multilocus Enzyme Electrophoresis
B. DNA Sequencing
C. Multilocus Sequence Typing
VII. The Organization of Genetic Diversity in Pathogenic Microorganisms
VIII. Population Genetics of Representative Bacterial Pathogens
A. Bordetella
B. Borrelia
C. Escherichia coli and Shigella
D. Haemophilus
E. Helicobacter
F. Listeria
G. Mycobacterium
H. Neisseria
I. Salmonella
J. Staphylococcus
K. Streptococcus
L. Vibrio
IX. Conclusions
References
I. Introduction
In what ways do pathogenic microorganisms differ from nonpathogenic forms? For an organism to be considered a pathogen, it must, during some phase of its life-cycle, advance disease and alter the health or behavior of another organism, that is, its host. Every organism can serve as a host for pathogens, and pathogens come into contact with a very large number of species; however, most pathogenic microorganisms are virulent to relatively few, and often only one, host species, and infection may cause disease in only a limited segment of the host population. So despite the wide range of mechanisms deployed by pathogens to disable their hosts (and to promote their own replication and transmission), there is one common theme: virulence depends upon the susceptibility of a host. Therefore, the identification of pathogens, the differences between pathogenic and nonpathogenic microorganisms, and the specific factors required for virulence must each be defined with regard to its relevance to the host.
This chapter addresses two general issues about the evolution of microbial pathogenesis. First, we consider the differences between the genomes of pathogenic and nonpathogenic bacteria, the specific types of genes that contribute to the virulence phenotype, and the evolutionary history of these sequences in the genome of pathogens. Next, we discuss the molecular population genetics of microbial pathogens and the factors that govern the organization of genetic diversity within these populations. Because the origins and genetic bases of virulence influence the population structure of pathogens, these topics are interconnected and broadly relevant to the emergence, outbreak, control, and prevention of the diseases caused by microbial pathogens.
II. The Genetic Basis of Virulence
One way to gain insight into the specific factors contributing to virulence has been to identify the genetic differences between pathogens and closely related nonpathogenic bacteria. In this regard, there are potentially four types of genetic events that might be responsible for the differences in pathogenic potential among related bacteria: (1) virulence as a result of genes specific to the pathogen; (2) virulence as a result of the absence of a suppressor locus in the pathogen; (3) virulence as a result of allelic differences between genes shared by the pathogen and nonpathogen; and (4) virulence as a result of the differential regulation of the same complement of genes in the pathogen and nonpathogen.
1. VIRULENCE AS A RESULT OF SPECIES- OR STRAIN-SPECIFIC GENES
The most common approach to investigate virulence genes—on both analytical and technical grounds—has been to search for sequences that are restricted to pathogenic organisms. Virulence determinants are often acquired through horizontal transfer, which may explain why virulence traits are distributed sporadically among bacterial taxa (Fig. 1) or within some bacterial species (Fig. 2).
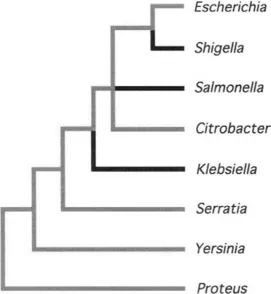
Fig. 1 Phylogenetic relationships among enteric bacteria showing taxa that are normally capable of invading eukaryotic cells (denoted with darkened branches).
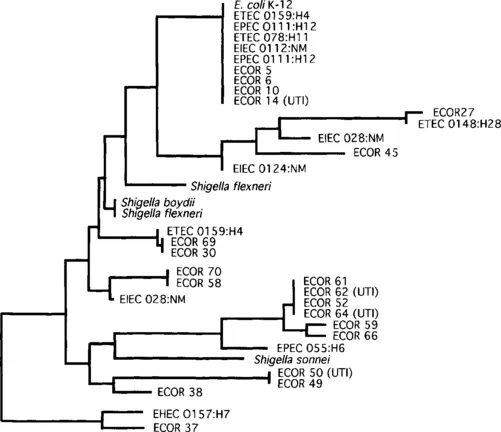
Fig. 2 Relationships among commensal and pathogenic strains of Escherichia coli and Shigella spp. based on nucleotide sequences of the gene encoding malate dehydrogenase. Abbreviations are as follows: UTI = urinary tract infection; EHEC = enterohemorrhagic E. coli; EIEC = enteroinvasive E. coli; EPEC = enteropathogenic E. coli; ETEC = enterotoxigenic E. coli. ECOR strains are from the E. coli reference collection [100], and the O:H serotypes of pathogenic E. coli are noted. (adapted with permission from Pupo et al. (1997) [56])
In many bacterial species, there is an association between species-specific genes and virulence. Pathogenicity islands (or Pai)—i.e., segments of the chromosome that encode virulence genes and are absent from related nonpathogenic bacteria [1–3]—have been identified in pathogenic strains of E. coli [4–6], Shigella flexneri [7, 8, 24], Salmonella enterica [9], Yersinia pestis [10–12], Vibrio cholerae [13], Haemophilus influenzae [14, 15], Helicobacter pylori [16], and Staphylococcus aureus [17]. In each of these cases, a specific chromosomally encoded gene, or cluster of genes, has been implicated in the virulence of the microorganism, and the corresponding region was not present in avirulent strains or related species.
Although it has long been known that species-specific regions confer traits that are unique to a bacterial species, the term “pathogenicity island” was first used to describe a DNA segment harbored by uropathogenic strains of E. coli [18]. Further characterization of pathogenicity islands revealed that many were situated at transfer RNA loci, which are commonly used as integration sites for foreign sequences [19]. For example, certain phages detected in E. coli, such as the retronphage ϕR73 [20] and the bacteriophage P4 [21] insert at, or near, tRNA genes, suggesting that pathogenicity islands are often transferred and acquired through phage-mediated events [1, 22]. The frequent insertion of foreign DNA sequences at tRNA genes is presumably due to the high degree of sequence conservation of tRNA genes across species. In fact, there is recurrent use of the tRNAselC locus, which is targeted by ϕR173 and as the integration site for several pathogenicity islands: Pai-1 of uropathogenic E. coli [4], the LEE island of enteropathogenic E. coli [23], the SHI-2 island of Shigella flexneri [8, 24], and the SPI-3 island of Salmonella enterica [25] each represent independent insertions of different virulence gene clusters into similar chromosomal locations. Flanking many pathogenicity islands there are signature sequences, such as short direct repeats, reminiscent of the integration of mobile elements (or even, in the case of Yersinia pestis, copies of the IS elements themselves) further attesting that these species- or strain-specific regions can be acquired laterally through a variety of transfer mechanisms.
Although research on pathogenicity islands focuses on chromosomally encoded regions, genes involved in bacterial virulence are also carried on extrachromosomal elements that are maintained within the genome of pathogens. For example, many of the genes required for Shigella virulence reside on a 220-kb plasmid [26, 27], and, similarly, all virulent strains of Yersinia harbor a 70- to 75-kb plasmid that encodes proteins necessary for their antihost properties [28, 29]. In this regard, the acquisition of plasmid-borne antibiotic resistance genes will also allow previously sequestered pathogens to exploit new hosts.
Other virulence determinants in these enteric species have been acquired by the organism in phage-mediated events. For example, the cytotoxins first characterized in Shigella are encoded on a bacteriophage that has subsequently been transferred to enterohemorrhagic strains of E. coli [30, 98]. In the case of Vibrio cholerae, two coordinately regulated factors contribute to virulence: cholera toxin, which is encoded by a filamentous bacteriophage (termed CTXϕ) related to the coliphage M13 [31], and the toxin-coregulated pili, which is encoded within a large pathogenicity island. This pathogenicity island of Vibrio cholera is in itself another filamentous bacteriophage [32], and thi...
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Contributors
- Preface
- Chapter 1: Evolution of Bacterial Pathogens
- Chapter 2: Germ Warfare: The Mechanisms of Virulence Factor Delivery
- Chapter 3: Regulation of Virulence Gene Expression in Bacterial Pathogens
- Chapter 4: Strategies to Identify Bacterial Pathogenicity Factors
- Chapter 5: Mechanisms of Bacterial Pathogenesis in Plants: Familiar Foes in a Foreign Kingdom
- Chapter 6: Yersinia
- Chapter 7: Molecular Pathogenesis of Salmonellae
- Chapter 8: Shigellosis: From Disease Symptoms to Molecular and Cellular Pathogenesis
- Chapter 9: Pathogenic Escherichia coli
- Chapter 10: Molecular Basis of Vibrio cholerae Pathogenesis
- Chapter 11: H. pylori Pathogenesis
- Chapter 12: Neisseria
- Chapter 13: Bordetella
- Chapter 14: Pathogenesis of Haemophilus influenzae Infections
- Chapter 15: Pathogenic Mechanisms in Streptococcal Diseases
- Chapter 16: Listeria monocytogenes
- Index
- Color Plates