
Mechanisms of Eukaryotic DNA Recombination
- 228 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Mechanisms of Eukaryotic DNA Recombination
About this book
Mechanisms of Eukaryotic DNA Recombination is a collection of papers that discusses advances in eukaryotic genetic recombination. Papers address issues in eukaryotic genetic recombination, particularly DNA integration in mammalian genomes, genetic recombination in Drosophila or Caenorhabditis; the manipulation of the mouse genome; genome organization; and genetic recombination in protozoa. One paper discusses chromatid interactions during intrachromosomal recombination in mammalian cells, namely, intrachromatid and sister chromatid. Another paper analyzes the implication for chromosomal recombination and gene targeting; results on extrachromosomal recombination show that circles are inefficient substrates for recombination even if only one of two substrates in an intermolecular reaction is circular. One author discusses the genetics and molecular biology of recombination, citing the work of Watson and Crick, stating that crossing-over occurs between genes (not within them). He also explains that the formation and resolution of recombination intermediaries depend on enzyme or other proteins. This book will prove invaluable to cellular biologists, microbiologists, and researchers engaged in genetics and general biology.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
Chromatid Interactions during Intrachromosomal Recombination in Mammalian Cells
Publisher Summary
INTRODUCTION
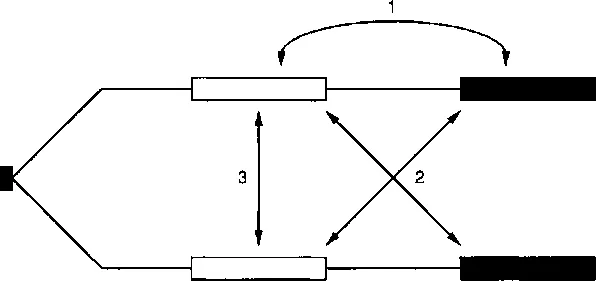
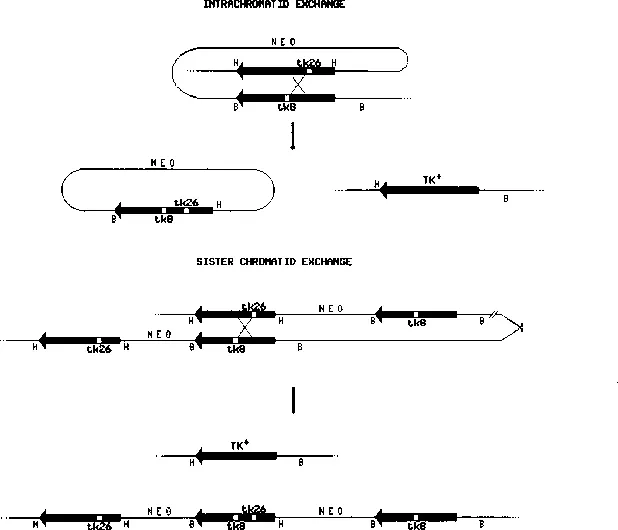
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Preface
- PART I: TARGETED DNA INTEGRATION IN MAMMALIAN CELLS
- PART II: DNA INSERTION PHENOMENA
- PART III: GENETIC RECOMBINATION IN DROSOPHILA AND CAENORHABDITIS
- PART IV: GENETIC RECOMBINATION IN YEASTS AND USTILAGO
- PART V: GENETIC RECOMBINATION IN TRYPANOSOMES AND PLASMODIUM
- Index