- 296 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Target Validation in Drug Discovery
About this book
This work presents a comprehensive contemporary framework for approaching target validation in drug discovery. It begins with a detailed description of new enabling technologies, including aptamers, RNA interference, functional genomics, and proteomics. The next section looks at biologic drug development with in-depth discussion of lessons learned from such well-known cases as Erbitux, Herceptin, and Avastin. Additional targets known as "second generation" drugs, which can be identified when disease pathways are validated by biologics, present new possible small molecule therapeutics and serve as the focus of the final section of the book.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Target Validation in Drug Discovery by Brian W. Metcalf,Susan Dillon in PDF and/or ePUB format, as well as other popular books in Medicine & Pharmacology. We have over one million books available in our catalogue for you to explore.
Information
Topic
MedicineSubtopic
PharmacologyIII
Validating targets of small molecule approaches
9
EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) INHIBITOR FOR ONCOLOGY: DISCOVERY AND DEVELOPMENT OF ERLOTINIB
KENNETH K. IWATA, Ph.D.*, SHANNON E. BEARD, Ph.D.† and JOHN D. HALEY*, *OSI Pharmaceuticals, Farmingdale, New York; †OSI Pharmaceuticals, Melville, New York
Molecular-targeted drug therapies are based upon our understanding of tumor-cell biology to identify oncogenic targets and develop therapeutic approaches to these targets using small-molecular-weight inhibitors, antibodies, and antisense. Unlike traditional chemotherapeutics, which rely upon differential toxicities to tumor cells versus normal cells, molecular-targeted therapy approaches identify key molecular drivers of tumorigenicity and develop drug therapies against the target. The goal of molecular-targeted therapies is to develop agents with better efficacy and tolerability than current chemotherapeutic drugs. During the past two decades, the increased understanding of the involvement of growth factors, receptors, and tyrosine kinases in tumor biology has made these oncogenes the focus of new, targeted cancer-drug-discovery efforts. The extensive literature on epidermal growth factor receptor (EGFR) and its ligands in tumor cell lines and clinical specimens has made EGFR an important molecular target for drug discovery. The focus of this chapter is to provide a background on EGFR as a cancer target, the development of erlotinib (a potent selective inhibitor of EGFR tyrosine kinase), and preclinical data supporting erlotinib’s advancement into the clinic. The concluding section will present some of the issues faced by molecular-targeted therapies such as erlotinib. These will include the role of biomarkers in the identification of patients who might best respond to treatment and efforts to optimize the use of molecular-targeted therapies.
I INTRODUCTION
Tyrosine kinases are among the earliest molecular targets identified for cancer-drug discovery (Gschwind et al., 2004). Determination that the DNA sequence for a virally associated oncogene v-sis has a proto-oncogene homologue c-sis, which encodes for the growth factor PDGFRβ (reviewed Heldin and Westermark, 1990), provided evidence that growth factors and growth factor receptors can drive tumorigenesis. The observation that growth factors, growth factor receptors, and kinases play a major role in oncogenesis, along with advances in molecular biology and automated high-throughput drug-screening technologies, has revolutionized approaches to small-molecule cancer-drug discovery resulting in the emergence of molecular targeted therapeutics. Unlike traditional chemotherapeutic drug discovery, which relies upon differential toxicities of normal cells versus tumor cells, targeted drug discovery identifies key molecular mediators of tumorigenicity against which small-molecule inhibitors are developed to generate cytostatic and/or cytotoxic effects on tumors with minimal negative effects on normal tissue and functions. Targeted approaches to anti-cancer therapy have the potential to further reduce toxicity and provide a better quality of life for the patient. EGFR and its ligands have long been observed to be elevated in many tumor cell lines (Arteaga, 2001; Salomon et al., 1995) and in tumor biopsy specimens and associated with poor clinical prognosis (Table 9.1). These observations made EGFR an ideal early target for molecularly targeted drug discovery.
TABLE 9.1
EGFR Over-Expression and Clinical Prognosis
| Tumor Type | Cases/Yeara | Percentb,c Over-expression | Related to Poor Prognosis | References |
| Prostate | 232,090 | 40–70 | Yes | Kim et al. 2001 |
| Scher et al. 1995 | ||||
| Breast | 212,930 | 35–70 | Yes | Nicholson et al. 2001 |
| NSCLC | 172,570 | 50–90 | Yes | Hirsch et al. 2003 |
| Selvaggi et al. 2004 | ||||
| Colon | 104,950 | 45–80 | Yes | Resnik et al. 2004 |
| Nicholson et al. 2001 | ||||
| O’Dwyer et al. 2002 | ||||
| Bladder | 63,210 | 30–90 | Yes | Neal et al. 1990 |
| Pancreatic | 32,180 | 30–50 | Yes | Xiong et al. 2002 |
| Papouchado et al. 2005 | ||||
| Head & Neck | 29,370 | 70–100 | Yes | Pomerantz and Grandis 2004 |
| Ovarian | 22,220 | 35–60 | Yes | Scambia et al. 1992 |
aAmerican Cancer Society 2005.
bCiardiello, F. et al. Expert Opin. Investig. Drugs (2002) 11: 755–768.
cSalomon, D. S. et al. Crit. Rev. Oncol. Hematol. (1995) 19: 183–232.
II EPIDERMAL GROWTH FACTOR RECEPTOR AND LIGANDS
Epidermal growth factor (EGF) is a highly conserved small molecular weight 6000 Dalton polypeptide that was first identified and purified by Stanley Cohen (reviewed Carpenter and Cohen, 1990).
EGFR is a 170-kDa transmembrane receptor tyrosine kinase. The EGFR gene encodes a 1186 amino acid polypeptide that is highly glycosylated in the extracellular ligand binding portion of the receptor. The extracellular domain consists of 621 amino acids including two cysteine-rich domains, to which N-linked carbohydrate is attached. The transmembrane domain contains 23 amino acids and the cytoplasmic domain, containing intrinsic tyrosine kinase activity, comprises of 542 amino acids.
Seven ligands for EGFR—EGF, TGFa, amphiregulin, betacellulin, epigen, epiregulin and heparin binding EGF-like growth factor (HB-EGF)—have been identified (Burgess et al., 2003). Crystallographic data suggest that the extracellular regions of EGFR function as a negative regulators of intrinsic kinase activity, which is relieved by interaction with ligand (Burgess et al., 2003, Ferguson et al., 2003, Schlessinger 2002). Binding of EGFR with ligand results in a conformational change, followed by receptor dimerization and activation of the tyrosine kinase domain. As shown in Figure 9.1, upon receptor activation, the EGFR transphosphorylates the tyrosine residues on the adjacent EGFR dimer.
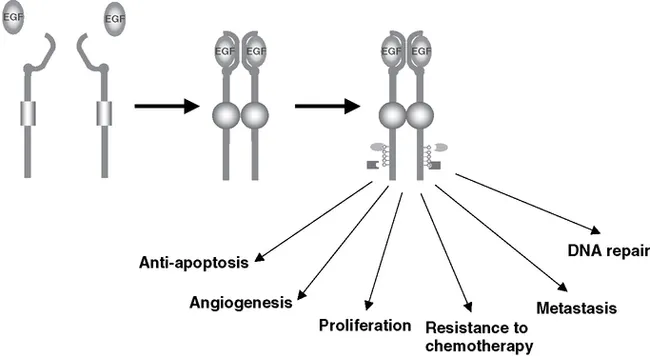
More than 12 tyrosines in the cytoplasmic domain of EGFR have been shown to be ...
Table of contents
- Cover image
- Title page
- Table of Contents
- PREFACE
- CONTRIBUTORS
- I: Pharmaceutical biotechnology for target validation
- II: Target validation for biopharmaceutical drug discovery
- III: Validating targets of small molecule approaches
- INDEX