
eBook - ePub
Metabolic Engineering
Principles and Methodologies
- 725 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Metabolic Engineering
Principles and Methodologies
About this book
Metabolic engineering is a new field with applications in the production of chemicals, fuels, materials, pharmaceuticals, and medicine at the genetic level. The field's novelty is in the synthesis of molecular biology techniques and the tools of mathematical analysis, which allow rational selection of targets for genetic modification through measurements and control of metabolic fluxes. The objective is to identify specific genetics or environmental manipulations that result in improvements in yield and productivities of biotechnological processes.
Key features of the book are pathway integration and the focus on metabolic flux as a fundamental determinant of cell physiology. The book keeps mathematical complexity to a minimum, and provides a glossary of biological terms to facilitate use of the book by a broader spectrum of readers. A web page exists to communicate updates of the codes and homework problems.
- Demonstrates metabolic engineering in action with numerous examples of pathway modification
- Includes methods for identifying key enzymes in metabolic networks
- Contains a comprehensive review of metabolic biochemistry
- Discusses metabolic regulation at the gene, enzyme, operon, and cell levels
- Explains concepts of stoichiometry, kinetics, and thermodynamics of metabolic pathways
- Minimizes mathematical complexity
- Links to a Web page to communicate updates of the software code and homework problems
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
CHAPTER 1 The Essence of Metabolic Engineering
The concept of metabolic pathway manipulation for the purpose of endowing microorganisms with desirable properties is a very old one indeed. We have many outstanding examples of this strategy in the areas of amino acids, antibiotics, solvents, and vitamin production. These methods rely heavily on the use of chemical mutagens and creative selection techniques to identify superior strains for achieving a certain objective. Despite widespread acceptance and impressive successes, the genetic and metabolic profiles of mutant strains were poorly characterized and mutagenesis remained a random process where science was complemented with elements of art.
The development of molecular biological techniques for deoxyribonucleic acid (DNA) recombination introduced a new dimension to pathway manipulation. Genetic engineering allowed the precise modification of specific enzymatic reaction(s) in metabolic pathways and, hence, the construction of well-defined genetic backgrounds. Shortly after the feasibility of recombinant DNA technology was established, various terms were coined to represent the potential applications of this technology to directed pathway modification. Some of the terms suggested were molecular breeding (Kellogg et al., 1981), in vitro evolution (Timmis et al., 1988), (microbial or metabolic) pathway engineering (MacQuitty, 1988; Tong et al., 1991), cellular engineering (Nerem, 1991), and metabolic engineering (Stephanopoulos and Vallino, 1991; Bailey, 1991). Although the exact definition varies from author to author, all convey similar meanings with respect to the general goals and means of metabolic engineering. Here we define metabolic engineering as the directed improvement of product formation or cellular properties through the modification of specific biochemical reaction(s) or the introduction of new one(s) with the use of recombinant DNA technology. An essential characteristic of the preceding definition is the specificity of the particular biochemical reaction(s) targeted for modification or to be newly introduced. Once such reaction targets are identified, established molecular biological techniques are applied in order to amplify, inhibit or delete, transfer, or deregulate the corresponding genes or enzymes. DNA recombination in a broader sense is routinely employed at various steps toward these ends.
Although a certain sense of direction is inherent in all strain improvement programs, the directionality of effort is a strong focal point of metabolic engineering compared to random mutagenesis, as it plays a dominant role in enzymatic target selection, experimental design, and data analysis. On the other hand, direction in cell improvement should not be interpreted as rational pathway design and modification, in the sense that it is totally decoupled from random mutagenesis. In fact, strains that are obtained by random mutation and exhibit superior properties can be the source of critical information about pathway configuration and control, extracted via reverse metabolic engineering.
As with all traditional fields of engineering, metabolic engineering too encompasses the two defining steps of analysis and synthesis. Because metabolic engineering emerged with DNA recombination as the enabling technology, attention initially was focused, almost exclusively, on the synthetic side of this field: expression of new genes in various host cells, amplification of endogenous enzymes, deletion of genes or modulation of enzymatic activity, transcriptional or enzymatic deregulation, etc. As such, metabolic engineering was, to a significant extent, the technological manifestation of applied molecular biology with very little engineering content. Bioprocess considerations do not qualify as metabolic engineering. A more significant engineering component can be found in the analytical side of metabolic engineering: How does one identify the important parameters that define the physiological state? How does one utilize this information to elucidate the control architecture of a metabolic network and then propose rational targets for modification to achieve a certain objective? How does one further assess the true biochemical impact of such genetic and enzymatic modifications in order to design the next round of pathway modifications and so on until the goal is attained? Instead of the mostly ad hoc target selection process, can one prescribe a rational process to identify the most promising targets for metabolic manipulation? These are some of the questions that the analytical side of metabolic engineering would address.
On the synthetic side, another novel aspect of metabolic engineering is the focus on integrated metabolic pathways instead of individual reactions. As such, it examines complete biochemical reaction networks, concerning itself with issues of pathway synthesis and thermodynamic feasibility, as well as pathway flux and its control. We thus are witnessing a paradigm shift away from individual enzymatic reactions and toward systems of interacting biochemical reactions. In this regard, the notion of the metabolic network is of central importance in the sense that an enhanced perspective of metabolism and cellular function can be obtained by considering a system of reactions in its entirety rather than reactions in isolation from one another. Through metabolic engineering, attention is shifted to the whole system instead of its constituent parts. In this regard, metabolic engineering seeks to synthesize and design using techniques and information developed from extensive reductionist research. In turn, observations about the behavior of the overall system are the best guide for further rational decomposition and analysis.
Although metabolism and cell physiology provide the main context for analyzing reaction pathways, it should be pointed out that results of flux determination and control have broader applicability. Thus, besides the analysis of material and energy fluxes through metabolic pathways, the concepts of metabolic engineering are equally applicable to the analysis of information fluxes as those encountered in signal transduction pathways. Because the latter have not yet been well-defined, the main focus of this book is on applications to metabolic pathways. However, once the concepts of information pathways have crystallized, we expect that many of the ideas and tools presented herein will find good use in the study of the interactions of signal transduction pathways and the elucidation of the complex mechanisms by which external stimuli control gene expression.
Perhaps the most significant contribution of metabolic engineering is the emphasis it places on metabolic fluxes and their control under in vivo conditions. To be sure, the concept of metabolic flux per se is not new. Metabolic flux and its control have occupied the attention of a small but forward thinking group of researchers in biochemistry for approximately 30 years. As a result of their work, ideas on metabolic control matured and were rigorously defined, although they were not always broadly embraced by traditional biochemists. Metabolic engineering, initially conceived as the ad hoc pathway manipulation, quickly became the natural outlet for the analytical skills of engineers who saw the opportunity for introducing rigor in this process by utilizing the available platform of metabolic control analysis. The combination of analytical methods to quantify fluxes and their control with molecular biological techniques to implement suggested genetic modifications is the essence of metabolic engineering. When practiced in an iterative manner, it provides a powerful method for the systematic improvement of cellular properties over a broad range of contexts and applications.
The flux is a fundamental determinant of cell physiology and the most critical parameter of a metabolic pathway. For the linear pathway of Fig. 1.1a, its flux, J, is equal to the rates of the individual reactions at steady state. Obviously, a steady state will be reached when the intermediate metabolites adjust to concentrations that make all reaction rates equal (v1 = v2 = … vi … = vL). During a transient, the individual reaction rates are not equal and pathway flux is variable and ill-defined (usually by the time varying rate of substrate uptake or product formation). For the branched pathway of Fig. 1.1b splitting at intermediate I, we have two additional fluxes for each of the branching pathways, related by J1 = J2 + J3 at steady state. The flux of each branch is equal to the rates of the individual reactions at the corresponding branch. It is often convenient to think of flux J1 as the superposition of the linear pathway fluxes J2 and J3, as shown in Fig. 1.1c. In this way, a complex network like the one depicted schematically in Fig. 1.1d can be decomposed into a number of linear pathways, each with its own flux as shown. It should be noted that, for all pathways of Fig. 1.1, a necessary condition to be able to reach steady state is that the rates of the initial and final reactions (or, equivalently, the concentrations of the initial and final metabolites, A and B, C, etc., respectively) must be constant. This is usually accomplished by constant extracellular metabolite concentrations in a continuous bioreactor, often referred to as a chemostat.
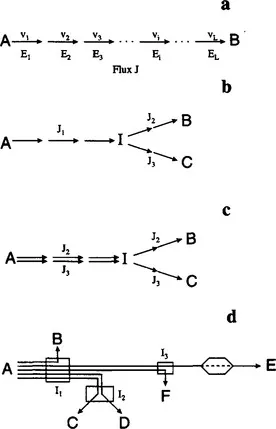
FIGURE 1.1 Examples of simple pathways.
As metabolic pathways and their fluxes are at the core of metabolic engineering, it is important to elaborate a little more on their definition and meaning. We define a metabolic pathway to be any sequence of feasible and observable biochemical reaction steps connecting a specified set of input and output metabolites. The pathway flux is then defined as the rate at which input metabolites are processed to form output metabolites. The importance of feasibility and observability should be noted. First, it would be of little value to create nonsense reaction sequences comprising enzymes that are not present in a cell. Similarly, no more valuable is the enumeration of feasible reaction sequences between substrates and products that, however, cannot be observed experimentally. This is a very important point in light of the diversity and complexity of the metabolic maps that have been constructed as result of pioneering research in biochemistry during the previous 50 years. Although there is often more than one bioreaction sequence between specified input and output metabolites, if the fluxes of these sequences cannot be determined independently, their inclusion provides no additional information. In many ways it is better if these reaction sequences are lumped together in fewer pathways whose fluxes can be observed. In the example of Fig. 1.1d, if the flux of each branch leading to the formation of metabolite E cannot be measured experimentally or otherwise determined, the two branches must be lumped into a single pathway shown by the dashed line. Clearly, the use of more informative measurements, such as those that are able to differentiate between the preceding two branches, should be encouraged as they enhance the resolution of biochemical pathways that can be observed through them. The determination of metabolic fluxes in vivo has been termed metabolic flux analysis (MFA) and is of central importance to metabolic engineering.
In this framework of metabolic pathways and fluxes, a fundamental objective of metabolic engineering is to elucidate the factors and mechanisms responsible for the control of metabolic flux. A better understanding of the control of flux provides the basis for rational modification of metabolic pathways. There are three steps in the process for the systematic investigation of metabolic fluxes and their control. The first is to develop the means to observe as many pathways as possible and to measure their fluxes. To this end, one starts with simple material balances based on the measurements of concentrations of extracellular metabolites. The measurement of metabolites A-F of the network of Fig. 1.1d allows one to determine the five indicated fluxes but not the pathway split before F. If, upon administration of a labeled precursor A, metabolite F is labeled differently when it is formed by a particular branch, then this method could provide information about the split flux ratio at the branch point before F. This is one of several techniques that can be applied to provide additional information about branching pathways, and they are discussed at considerable length in this book. It is essential to emphasize that the flux of a metabolic pathway is not the same as the enzymatic activity of one or more of the enzymes in the pathway. In fact, enzymatic assays provide no information about the actual flux of the pathway other than that the corresponding enzyme is present and active under the in vitro assay conditions. Their inclusion in metabolic studies has often been misinterpreted to imply a metabolic flux of similar magnitude, which is certainly incorrect to generally conclude.
The second step is to introduce well-defined perturbations to the bioreaction network and to determine the pathway fluxes after the system relaxes to its new steady state. Because all flux control investigations quickly focus on a particular metabolic branch point, it is convenient to think of flux control in terms of the schematic branched pathway of Fig. 1.2. Three perturbations are required, in general, to study this branch point, each one originating in each of the corresponding branches. The ideal perturbation would involve a chemostat, where the activity of an enzyme is suddenly perturbed (through, for example, the use of an inducible promoter) after the system has reached a steady state. This arrangement is most applicable to microbial and cell culture systems. Different experimental configurations will be needed for other types of systems such as plants and organ function in vivo. Other types of perturbations that are easier to implement, such as the addition of a pulse of substrate or switching to a different carbon source, can be also informative. Perturbations should be targeted toward enzymes close to the branch point, although any other perturbation with an appreciable effect on the corresponding branch flux should be acceptable. Finally, one should note that one perturbation can provide information about more than one branch point, and this is useful for minimizing the number of experiments required to elucidate the control structure of a realistic metabolic network.
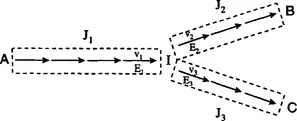
FIGURE 1.2 Branched pathway where the individual reactions are grouped.
The third and final step in flux control determination is analysis of the flux perturbation results. Clearly, perturbation of the flux of each of the three branches of Fig. 1.2 allows one to probe the flexibility of the particular branch point. For example, if a large perturbation of flux J1 has no appreciable effect on the magnitude ...
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Dedication
- PREFACE
- LIST OF SYMBOLS
- Chapter 1: The Essence of Metabolic Engineering
- Chapter 2: Review of Cellular Metabolism
- Chapter 3: Comprehensive Models for Cellular Reactions
- Chapter 4: Material Balances and Data Consistency
- Chapter 5: Regulation of Metabolic Pathways
- Chapter 6: Examples of Pathway Manipulations: Metabolic Engineering in Practice
- Chapter 7: Metabolic Pathway Synthesis
- Chapter 8: Metabolic Flux Analysis
- Chapter 9: Methods for the Experimental Determination of Metabolic Fluxes by Isotope Labeling
- Chapter 10: Applications of Metabolic Flux Analysis
- Chapter 11: Metabolic Control Analysis
- Chapter 12: Analysis of Structure of Metabolic Networks
- Chapter 13: Flux Analysis of Metabolic Networks
- Chapter 14: Thermodynamics of Cellular Processes
- GLOSSARY
- INDEX
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access Metabolic Engineering by George Stephanopoulos,Aristos A. Aristidou,Jens Nielsen in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Medical Theory, Practice & Reference. We have over one million books available in our catalogue for you to explore.