
- 396 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Genetical Analysis of Quantitative Traits
About this book
This text provides a guide to the experimental and analytical methodologies available to study quantitative traits, a review of the genetic control of quantitative traits, and a discussion of how this knowledge can be applied to breeding problems and evolution.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Genetical Analysis of Quantitative Traits by Dr M Kearsey,Dr H Pooni in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.
Information




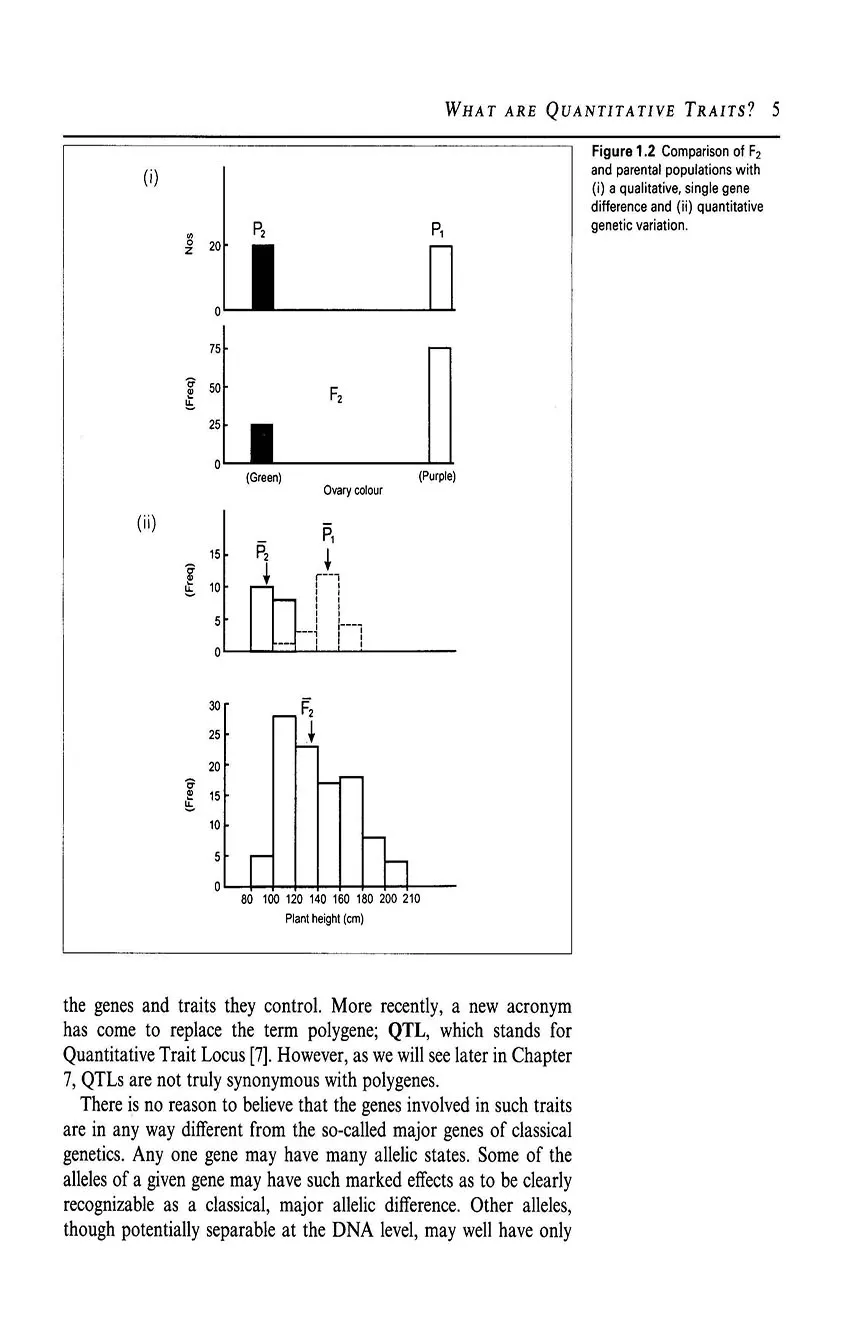



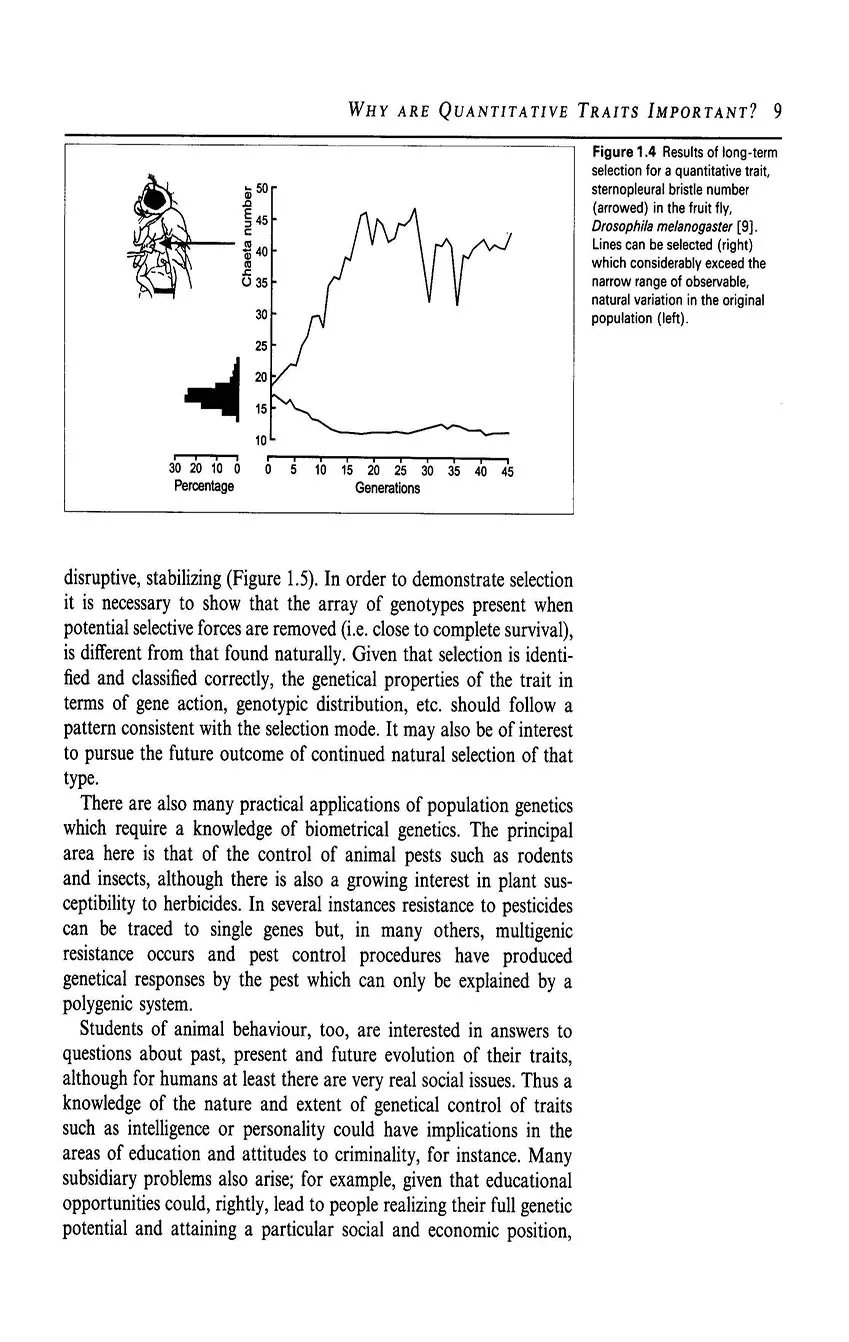
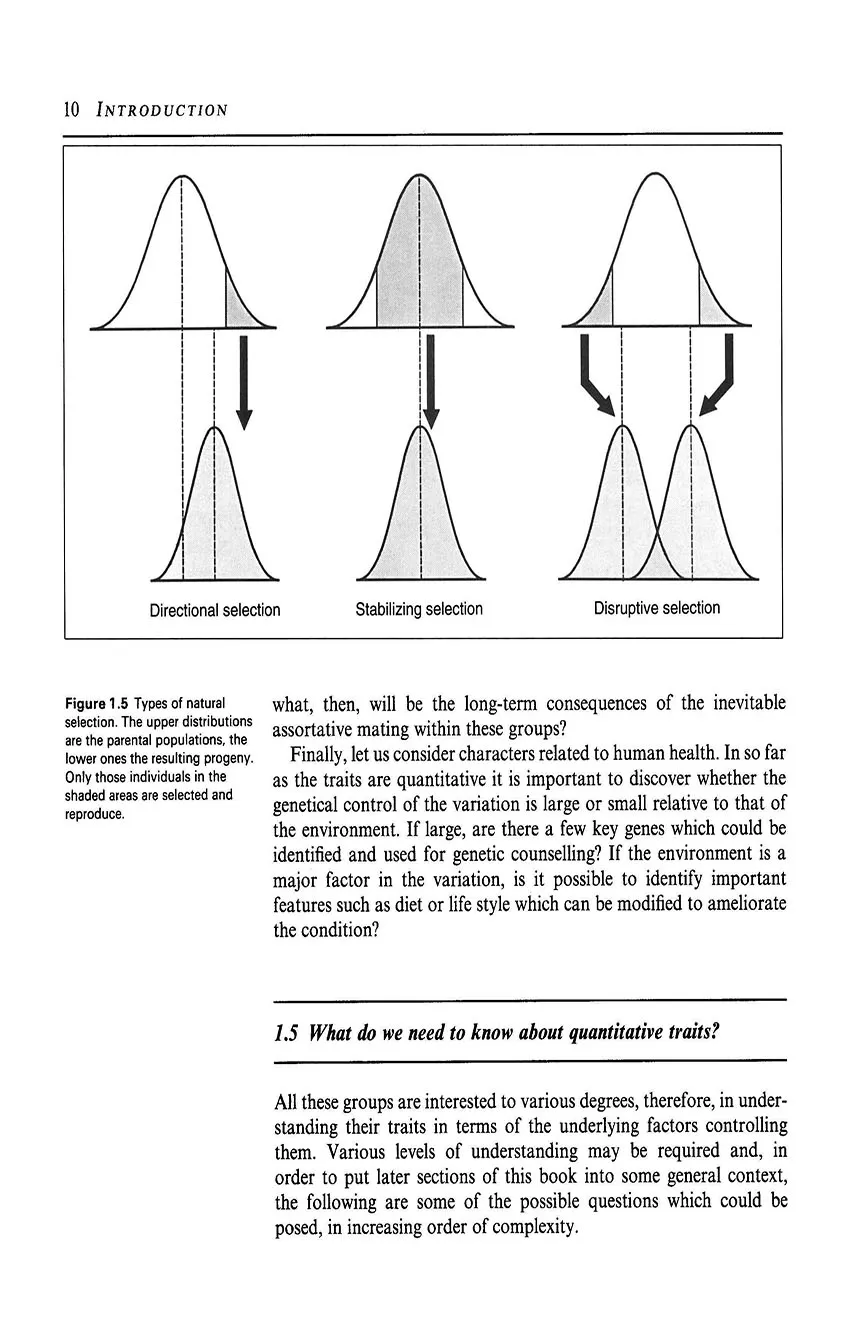












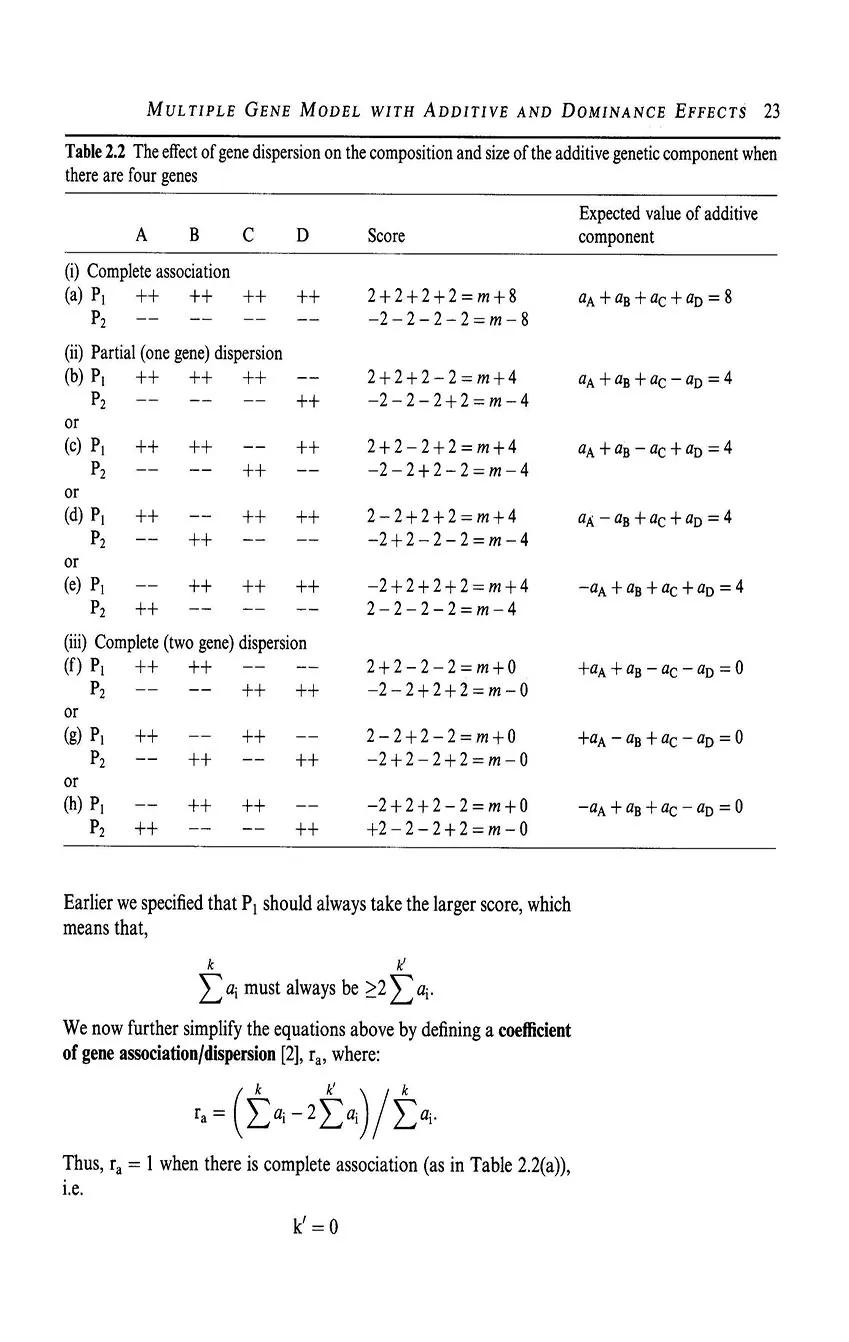


Table of contents
- Cover
- Half Title
- Title Page
- Copyright Page
- Contents
- Preface
- Dedication
- 1 Introduction
- 1.1 Qualitative, single gene differences
- 1.2 What are quantitative traits?
- 1.3 Who studies quantitative traits?
- 1.4 Why are quantitative traits important?
- 1.5 What do we need to know about quantitative traits?
- 1.6 Historical development of quantitative genetics
- Summary
- Further reading
- 2 Basic generations - means
- 2.1 Single gene model with additive and dominance effects
- 2.2 Two gene model with additive and dominance effects
- 2.3 Multiple gene model with additive and dominance effects
- 2.4 Extension to other generations
- 2.5 Relationships between generation means
- 2.6 Estimating genetical parameters
- 2.7 Interpretation: heterosis and potence ratio
- Summary
- References
- 3 Basic generations- variances
- 3.2 Environmental variation
- 3.3 Estimating environmental variance
- 3.4 Variation in the segregating generations
- 3.5 Estimation of genetical components
- 3.6 Heritability, h2
- 3.7 Relationships between [a] and V*a, and [d] and V*D
- 3.8 Dominance ratio
- 3.9 Variances and means
- References
- 4 Selfing and full-sib mating
- 4.1 Selfing: F3 families
- 4.2 Selfing: F4 families and beyond
- 4.3 Applications of selfing theory
- 4.4 Variation between inbred lines derived from an F2
- 4.5 Sib-mating an F2
- 4.6 Sib-mating: regression of offspring onto the parents
- 4.7 Further inbreeding by full-sib mating
- 4.8 Estimation of variance components by weighted least squares
- 4.9 Unequal family sizes
- Summary
- References
- 5 Half-sib mating designs
- 5.1 The North Carolina Experiment I: NCI
- 5.2 The North Carolina Experiment II: NCII
- 5.3 General and specific combining ability
- 5.4 Multiple NCIIs
- 5.5 The North Carolina Experiment III: NCIII
- 5.6 The Triple Test Cross: TTC
- 5.7 The diallel cross
- 5.8 HS designs using inbred lines from an F2 as parents
- Summary
- References
- 6 Genes, genetic markers and maps
- 6.1 Genetic markers
- 6.3 Mutations in structural genes
- 6.4 Molecular genetic markers
- 6.5 Chiasmata, crossing over and genetic exchange
- 6.6 Chiasma frequency and recombination frequency
- 6.7 Estimation of recombination frequency
- 6.8 Mapping functions
- 6.9 Segregation distortion
- Summary
- References
- Further reading
- 7.2 Background to methodology
- 7.3 QTL and marker loci in segregating generations
- 7.4 Handling more than one QTL on a chromosome
- 7.5 Biometrical methods of gene counting
- 7.6 General conclusions on QTL counting and locating
- Summary
- References
- Further reading
- 8 Designer chromosomes
- 8.1 Chromosome engineering
- 8.2 Locating a QTL within the substituted region
- 8.3 Detecting substitution effects
- 8.4 Manipulating whole chromosomes
- 8.5 Chromosome substitution methods in different species
- 8.6 Use of chromosome substitution lines
- Summary
- References
- 9 Populations
- 9.2 Solutions
- 9.3 Consequences of definitions of VA and VD
- 9.4 Studies of human populations
- 9.5 The use of twins
- 9.6 Heritabilities of human traits
- 9.7 Genotype-environment correlation
- 9.8 Diallel crosses
- 9.9 Inbreeding in a population
- Summary
- References
- 10 The consequences of linkage
- 10.1 Genetic variation with linkage
- 10.2 Extension to more than two genes
- 10.3 Linkage and random mating an F2
- 10.4 Linkage and inbreeding an F2
- 10.6 Tests of linkage
- 10.7 Sex linkage
- 10.8 Basic generations of single crosses
- 10.9 Full-sib families
- 10.10 Half-sib designs
- Summary
- References
- 11 Epistasis
- 11.1 Definitions
- 11.2 Relationship with classical epistasis
- 11.3 What is parameter m?
- 11.4 The effects of association and dispersion on epistasis
- 11.5 Deriving the expectations of generation means
- 11.6 Estimates and tests of significance
- 11.7 Determining the type of epistasis for a multigene case
- 11.9 Higher order interactions
- 11.10 Epistasis and variances
- Summary
- References
- 12 Genotype by environment interaction
- 12.1 The nature and causes of G x E
- 12.2 Tests of G x E
- 12.3 Macro-environ-mental variables
- 12.4 G x E with many lines
- 12.5 Interpretation of G x E analysis
- 12.6 Predictions in the presence of G x E
- 12.7 Conclusions from the genetic analysis of G x E
- 12.8 Selection in heterogeneous environments
- 12.9 Other methods of analysis
- 12.10 G x E- stability versus flexibility
- Summary
- References
- Further reading
- 13 Maternal effects and non-diploids
- 13.2 Models for generation means
- 13.3 Maternal effects and scaling tests
- 13.4 Generation variances
- 13.5 Maternal effects in FS and HS designs
- 13.6 Haploids and polyploids
- 13.7 Basic generations with haploids
- 13.8 Multiple mating designs with haploids
- 13.9 Basic generations with polyploids
- 13.10 Multiple mating designs with polyploids
- Summary
- References
- 14 Correlated and threshold characters
- 14.1 Correlations between characters
- 14.2 Environmental correlations
- 14.3 Genetic correlations
- 14.4 Genetic covariation and design of experiment
- 14.5 Causes of covariation
- 14.6 General conclusions about correlations
- 14.7 Threshold traits
- 14.8 Handling threshold traits
- 14.9 Two or more thresholds
- Summary
- Further reading
- 15 Applications
- 15.1 Choice of breeding objective
- 15.2 The causes of heterosis
- 15.3 Predicting the breeding potential of crosses
- 15.4 Effects of failed assumptions on predictions of RILsand SCHs
- 15.5 Predicting the response to selection
- 15.6 Correlated response to selection
- 15.7 Indirect selection
- 15.8 Multi-trait selection
- 15.9 Marker-based selection
- 15.10 Genetic architecture of populations
- 15.11 Human populations
- Summary
- References
- Further reading
- 16 Experimental design
- 16.1 Replication
- 16.2 Power of experiments
- 16.3 Power of biometrical experiments ba,sed on ANOVA
- 16.4 Reliability of the additive genetic variance
- 16.5 Data analysis
- Summary
- References
- Appendix A Precision of h2n with FS families
- Appendix B Precision of h2n with HS families
- Appendix C Statistical tables; F, χ2, t
- Appendix D Normal deviate and intensity of selection (i)
- Appendix E Area under the normal curve
- Appendix F The weighted least squares procedure
- Symbols
- Problems
- Answers to problems
- Index