eBook - ePub
Affinity Chromatography
Template Chromatography of Nucleic Acids and Proteins
- 248 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
About this book
This book informs the reader about the practical methods, possibilities, and limits of template chromatography. It shows the various techniques for immobilization of nucleic acids fragments, polynucleotides, and nucleic acids by which the desired separation of materials can be achieved.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Affinity Chromatography by Herbert Schott in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.
Information
1
Introduction
It is known that the exact replication of genetic information is controlled by the base pair mechanism described by Watson and Crick. This mechanism depends on the fact that Ade and Cyt undergo specific interaction with Thy and Gua, respectively.
The base pairs Thy ·Ade and Cyt ·Gua are thus referred to as complementary base pairs. Single strands of nucleic acids posessing complementary base sequences will aggregate to double strands under certain conditions. The association is not restricted to complementary single strands of nucleic acids but can, under certain conditions, also be observed among poly-, oligo-, and mono-nucleotides; nucleosides, and nucleobases.
Since base pairing is specific and reversible, it recommends itself as the basis of a form of affinity chromatography suitable for the separation and isolation of nucleic acids and their fragments. This is accomplished in practice by immobilizing defined nucleic acids, polynucleotides, oligonucleotides, or nucleic acid residues on a stationary phase. If the conditions of base pairing are fulfilled, the complementary partner in the mobile phase is strongly adsorbed by the stationary complementary partner, non-complementary partners being retarded to only a small extent or not at all. After the elution of the noncomplementary components, the conditions of elution are altered such that base pairing is eliminated, and the adsorbed compounds are desorbed.
These immobilized compounds have the same template function as DNA, since only the nucleotide sequence of the immobilized compound determines which partner is complementary and which is thus adsorbed. The template function can be used additionally for the isolation of enzymes and other natural substances if they are capable of undergoing specific interactions with stationary nucleotide sequences.
This special form of affinity chromatography, which has shown rapid development in the last few years, may thus be termed template chromatography. The method has established itself as a useful technique which has recently received diverse applications in molecular biology as well as in the study of enzymes. The use of template chromatography in the study of enzymes is not surprising since about 30% of the approximately 2000 enzymes found in the cell participate in reactions involving nucleotides/nucleotide coenzymes.
The requirements of a suitable system for template chromatography are dictated by the moiety to be purified, the coupling of ligand to matrix, and the working milieu. For many research purposes details of molecular parameters may be vital: knowledge and reproducibility of coupling modes, matrix characteristics, and ligand-matrix-medium interactions then assume paramount importance. For large-scale production in particular, mechanical stability, rapid flow rate, resistance to microbial attack, and reuse are important factors. It will often be necessary to space the ligand from its support. This may be achieved either by coupling the ligand to one end of an “arm,” the other end of which is subsequently attached to the carrier, or by coupling it to an arm already modifying the matrix. This precaution will be superfluous if the ligand is bulky enough to make its binding site(s) available.
The basic problem in template chromatography, that of immobilizing the nucleic acids and their fragments on a suitable support in order to form the stationary phase, may be solved in several ways, as will be shown.
2
Immobilization of Nucleoside Phosphates, Nucleosides, and Nucleobases
It is the aim of this chapter to review the basic strategies of immobilizing nucleoside phosphates, nucleosides, and nucleobases to insoluble supports. The affinity between the covalently bound nucleic acid component and the partner of the mobile phase to be isolated is highly stereospecific. The interaction does not only lead to a complex formation. The subsequent displacement of the molecule from the complex will depend (a) on the three-dimensional orientation, (b) on the group and position on the ligand molecule through which it is immobilized to the support, (c) to some extent on the chemical method by which the column is made, and (d) on the spacer distance between the ligand and the supporting matrix. Moreover, the exact mechanism by which the immobilized ligand interacts with the partner of the mobile phase is very important for the design of the optimal matrix.
Columns with immobilized nucleic acid components were prepared particularly with a view to using them to investigate the wide field of enzymes that interact with nucleic acids and/or their components. The variety of these enzymes required template columns in which different positions of the nucleic acid components are bound to polymer support.
I. IMMOBILIZATION OF NUCLEOSIDE PHOSPHATES
Figure 1, for example, demonstrates the possible positions on AMP that can be functionalized for the immobilization to a polymer support. In essence, these can be classified into (a) phosphate-linked and (b) sugar-linked derivatives having an unsubstituted nucleobase, and (c) base-linked nucleoside phosphates containing free sugar and phosphate groups.
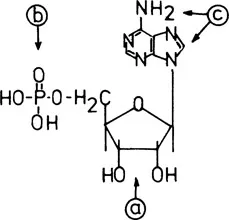
Figure 1 Positions of AMP that can be derivatized: (a) ribosyl-linked; (b) phosphate-linked; (e) N6-linked and C-8-linked. (From Ref. 3.)
An immobilization of nucleotides in which several positions of the ligand are simultaneously involved, for example, amide linkage through an amino group, esterification of sugar hydroxyls, or anhydride formation with the phosphoric acid residue, results in an ill-defined matrix. Rather, it is better to aim for methods that result in a more defined matrix, the interaction of which can be more exactly predicted and controlled.
A lot of literature has been concerned with designing simplified chemical methods for preparing polymer support derivatized with nucleic acid components. The immobilization of ligands to several supports (e.g., CNBr-Sepharose, AE-Sepharose, CM-eellulose, AE-cellulose, and functionalized nylon) is described [1] and the yields of several reactions are summarized in Table 1.
| Coupling yields | ||
|---|---|---|
| Ligand | ymol | % |
| | ||
| Coupling of ligands to 500 mg of Sepharose by the CNBr method: | ||
| p-Aminophenyl ATP | 20 | |
| 6-Aminohexanoyl ATP | 90 | |
| s6 ITP | 43 | |
| Coupling of ligand to 500 mg of CM-eellulose by the mixed carbonic anhydride method: | ||
| ATP | 6.0 | 16 |
| Coupling of halogen-substituted ligands to 200 mg of AE-cellulose: | ||
| br8 ATP | 2.0 | 40 |
| cl6 ITP | 24 | 72 |
| Coupling of ligand to 500 mg of AE-Sepharose by the glutaraldehyde method: | ||
| ATP | 4.1 | 20 |
| Coupling of ligands to 500 mg of AE-cellulose by bisimidate cross-linking: | ||
| AMP | 2.8 | 20 |
| ATP | 2.5 | 22 |
| Coupling of ligand to 200 mg of nylon 6 powder by the methylation procedure: | ||
| ATP | 4.0 | 80 |
Source: Data from Ref. 1.
The most exploited procedure for immobilization of nucleic acid components and their derivatives is the CNBr method, which has been reported by many laboratories. The results described, however, are very different [3–6]. The major problem in the preparation of adsorbents suitable for template chromatography is the design and synthesis of nucleotide ligands to be attached to the insoluble support.
A. Phosphate–Linked Nucleoside Phosphates
Linkage through the phosphat...
Table of contents
- Cover
- Half Page
- Series Page
- Title Page
- Copyright Page
- Preface
- Contents
- Abbreviations
- 1 Introduction
- 2 Immobilization of Nucleoside Phosphates, Nucleosides, and Nucleobases
- 3 Immobilization of Oligonucleotides
- 4 Immobilization of Polynucleotides and Nucleic Acids
- 5 Chromatography of Oligonucleotides
- 6 Isolation of DNA and its Fragments
- 7 Isolation of RNA
- 8 Isolation of Messenger RNA
- 9 Isolation of Proteins
- 10 Enzymic Synthesis or Degradation of Polynucleotides Using Polymer-Bound Primers, Templates, or Substrates
- 11 Studies of Peptide-Nucleotide Interactions
- References
- Index