
Metagenomics and Microbial Ecology
Techniques and Applications
- 200 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Metagenomics and Microbial Ecology
Techniques and Applications
About this book
Microorganisms comprise the greatest genetic diversity in the natural ecosystem, and characterization of these microbes is an essential step towards discovering novel products or understanding complex biological mechanisms. The advancement of metagenomics coupled with the introduction of high-throughput, cost-effective NGS technology has expanded the possibilities of microbial research in various biological systems. In addition to traditional culture and biochemical characteristics, omics approaches (metagenomics, metaproteomics, and metatranscriptomics) are useful for analyzing complete microbial communities and their functional attributes in various environments.
Metagenomics and Microbial Ecology: Techniques and Applications explores the most recent advances in metagenomics research in the landscape of next-generation sequencing technologies. This book also describes how advances in sequencing technologies are used to study invisible microbes as well as the relationships between microorganisms in their respective environments.
Features:
- Covers a wide range of concepts, investigations, and technological advancement in metagenomics at the global level.
- Highlights the novel and recent approaches to analyze microbial diversity and its functional attributes.
- Features a range of chapters that present an introduction to the field and functional insight into various ecosystems.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
Section IV
Metagenomics of Various Ecotypes
6
Earthworm Gut Microbiome
The Uncharted Microbiome
CONTENTS
6.1 Introduction
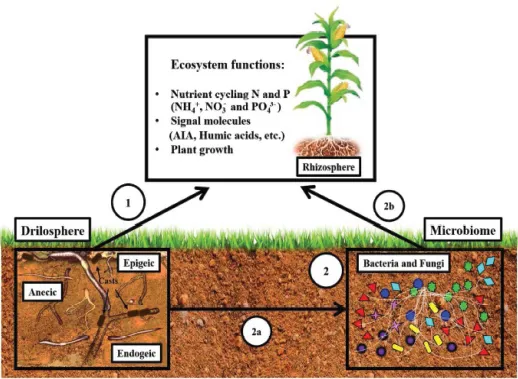
6.2 Microbiome of Earthworm
| Microbe Name | Family | Function | Reference |
|---|---|---|---|
| Dermacoccus | Dermacoccaceae | Heavy metal resistant (cadmium) | Šrut et al. 2019 |
| Rathayibacter | Microbacteriaceae | ||
| Sanguibacter | Sanguibacteraceae | ||
| Chryseobacterium | Flavobacteriaceae | ||
| Solibacillus | Planococcaceae | ||
| Cohnella | Paenibacillaceae | Cellulolytic bacterial | |
| Streptomyces | Streptomycetaceae | Heavy metal resistant (cadmium); glucose isomerase activities | Thakuria et al. 2010; Šrut et al. 2019 |
| Rhizobium | Rhizobiaceae | Bio-fertilizer | Hussain et al. 2016; Šrut et al. 2019 |
| Symbiobacterium | Symbiobacteriaceae | NS | |
| Rhodobacter | Rhodobacteraceae | Perform anoxygenic photosynthesis, degrade insecticide (Acephate) | Drake and Horn 2007; Phugare et al. 2012; Šrut et al. 2019 |
| Paenibacillus sp. | Paenibacillaceae | N2O-producing | Ihssen et al. 2003; Šrut et al. 2019 |
| Protochlamydia/Candidatus | Parachlamydiaceae | Symbionts of earthworms, heavy metal resistant (cadmium) | Nechitaylo et al. 2009; Šrut et al. 2019 |
| Rummeliibacillus | Planococcaceae/Caryophanaceae | NS | Šrut et al. 2019 |
| Shewanella | Shewanellaceae | NS | |
| Pseudomonas sp. | Pseudomonadaceae | NO2-producing | Drake and Horn 2007; Ihssen et al. 2003; Šrut et al. 2019 |
| Roseococcus sp. | Acetobacteraceae | Endosulfan-degrading; chlorinated hydrocarbon-degrading | Šrut et al. 2019; Thakuria et al. 2010; Verma et al. 2006 |
| Patulibacter | Patulibacteraceae | NS | Šrut et al. 2019 |
| Yersinia | Enterobacteriaceae | NS | |
| Parvibaculum | Rhodobiaceae | NS | |
| Agrobacterium | Rhizobiaceae | Heavy metal resistant (cadmium) | |
| Ochrobactrum | Brucellaceae | NS | |
| Thermoactinomyces | Thermoactinomycetaceae | NS | |
| Exiguobacterium | Bacillaceae | Degrade... |
Table of contents
- Cover
- Half Title Page
- Title Page
- Copyright Page
- Contents
- Preface
- Editors
- Contributors
- Section I An Overview of Metagenomics
- Section II Metagenomics Tools to Access Microbial Diversity
- Section III Metagenomics of Extreme Environments
- Section IV Metagenomics of Various Ecotypes
- Section V Applications
- Index