
- 328 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Programmed Cell Death in Animals and Plants
About this book
This title, the 52nd in symposium series, presents a comprehensive review of apoptosis and features contributions from many internationally recognised authors. The book is the first to integrate programmed cell death in plants, invertebrates and vertebrates. Rapid publication ensures that the very latest research is included.
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription.
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn more here.
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes! You can use the Perlego app on both iOS or Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Yes, you can access Programmed Cell Death in Animals and Plants by John Bryant,Jeff Garland,S.G. Hughes in PDF and/or ePUB format, as well as other popular books in Scienze biologiche & Biologia. We have over one million books available in our catalogue for you to explore.
Information
Topic
Scienze biologicheSubtopic
Biologia1 Apoptotic cell death: from worms to wombats … but what about the weeds?
DOI: 10.1201/9781003076889-1
1. Caenorhabditis elegans: the worm that turned … into a model multicellular eukaryote
Caenorhabditis elegans is a nematode (roundworm) whose potential for use as a model for genetic regulation of development was first recognized by Sydney Brenner as long ago as the 1960s (Brenner, 1974; Wood, 1988). The hermaphrodite worms can be grown easily in culture on agar in petri dishes, feeding on E. coli (model eats model!). The adult hermaphrodite is about 1 mm long (Figure 1) and has exactly 959 somatic cells (so it is certainly a very simple metazoan); the origins, lineages and developmental pathways of each of these cells have been precisely described (Sulston and Horvitz, 1977; Kimble and Hirsch, 1979; Sulston et al., 1983). The relevance of those developmental pathways for this book is that 1090 somatic cells are actually generated during development; the final number of 959 is achieved by the elimination via programmed cell death of 131 specific cells. Thus, programmed cell death is a normal part of development and because it always occurs in the same place and at the same stage of development, the process is readily studied.
As research on development of C. elegans progressed, mutants were discovered that had very specific effects on cell fate and function (as discussed in respect of cell death in subsequent sections of this chapter). The obvious usefulness of the genetic approach to studying development, taken with the relatively small genome size of C. elegans, led to its inclusion as one of the specific targets for genome sequencing. The first eukaryotic genome to be sequenced was that of budding yeast, Saccharomyces cerevisiae, completed in 1996 (Goffeau et al., 1996). The full genome sequence of its very distant cousin, the fission yeast, Schizosaccharomyces pombe, is likely to be known as this book is published in early 2000. However, between those two events, and very much to the credit of the C. elegans sequencing consortium, the essentially complete sequence of the genome of what is affectionately known as ‘the worm’, was known at the end of 1998 (The C. elegans Sequencing Consortium, 1998; Hodgkin et al., 1998). This was a very significant event, being the first complete elucidation of the genome sequence of a multicellular organism. The genome consists of 97 × 106 base pairs (about eight times the size of the S. cerevisiae genome and just under one thirtieth the size of the human genome). These 97 million base pairs make up about 19 000 predicted protein-coding genes (humans have nearly five times as many), over 40% of which have significant matches with genes in other organisms. The genes that regulate programmed cell death in C. elegans are within this 40%. The value of this organism in understanding the essential features of cell death is thus apparent.
2. The molecular mechanisms of programmed cell death in Caenorhabditis elegans
The detailed knowledge of the location and timing of programmed cell death during development of the adult hermaphrodite C. elegans has led in turn to screening for mutations that affect particular steps in the regulation and actuation of the cell death pathway. Analysis of mutant phenotypes suggests that the genes identified in the mutant screens fall into four general groups (Figure 2; Hengartner and Horvitz, 1994b; Hengartner, 1999) which, in order of action, are as follows:
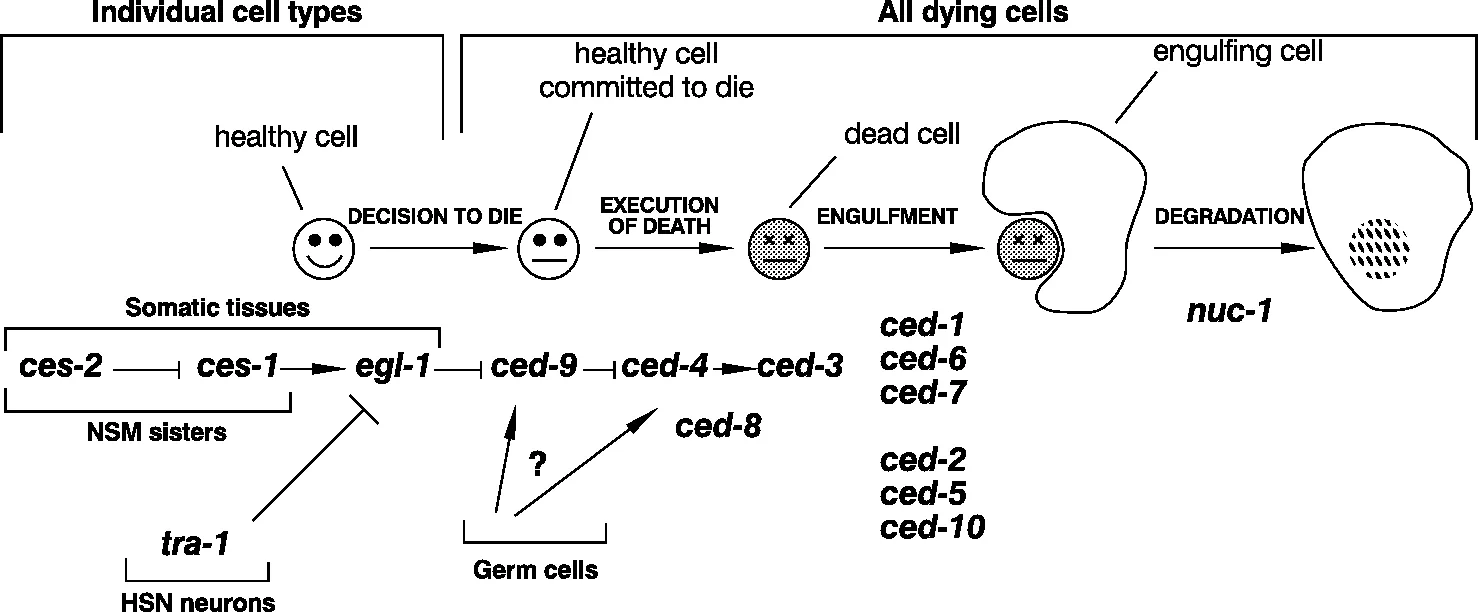
- There are several genes involved in the selection of the particular cell types that are to die, including ces-1 and -2 (ces = cell death specification).
- Five genes control the actual killing of the selected cells. These are ced-3, -4, -8, -9 and egl-1 (ced = cell death abnormal and egl = egg-laying defective).
- Six genes code for proteins required for the recognition, engulfment and removal of the dying cells. Functionally they fall into two groups, ced-1, -6 and -7 and ced-2, -5, and -10.
- The product of the nuc-1 (nuc = nuclease) gene completes degradation of the genomic DNA of dead cells.
2.1 The ‘killer genes’ and their regulation
Two genes, ced-3 and ced-4 are both absolutely required for essentially all programmed cell deaths in C. elegans. Mutational inactivation of either of these genes leads to the survival of all the 131 cells that normally die during development of the hermaphrodite adult (Ellis and Horwitz, 1986). A key question here is whether the products of these genes act from outside or within the cells that are killed, i.e., are the dying cells ‘murdered’ or do they ‘commit suicide’? The answer comes from mosaic analysis and from recent experiments using ectopic expression that clearly show that both these genes act within the dying cells themselves and thus the cells bring about their own death (Yuan and Horvitz, 1990; Shaham and Horvitz, 1996).
If ced-3 and ced-4 may be called ‘killer’ genes then ced-9 is a ‘moderator’ or ‘regulator’ gene (Hengartner et al., 1992). In ced-9 mutants exhibiting loss-of-function, many cells that should survive instead become apoptotic and die. In contrast to this, transgenic worms that over-express ced-9 or worms with ced-9 mutations causing gain-of-function fail to activate the apoptosis programme and so cells that should die do not. These data suggest that the normal role of ced-9 is to prevent the inappropriate action of ced-3 and ced-4 via an interaction of the CED-9 protein with the CED-3 and/or CED-4 proteins. In fact, interaction with the latter is strongly indicated by several lines of evidence: genetic analysis suggests that the ced-4 gene acts upstream of the ced-3 gene (Shaham and Horvitz, 1996). Interaction between the CED-9 and the CED-4 proteins has been demonstrated directly in vitro and in the yeast two-hybrid system (Chinnaiyan et al., 1997b; Irmler et al., 1997; James et al., 1997; Ottilie et al., 1997; Wu et al., 1997), whilst mutation analysis has identified the domains in CED-9 that facilitate the interactions with CED-4 (Spector et al., 1997). CED-4 also interacts with the pro-form of the CED-3 caspase (a caspase is an apoptotic cysteine protease); this interaction leads indirectly to the processing of pro-CED-3 and hence the production of active CED-3 (Chinnaiyan et al., 1997a; Seshagiri and Miller, 1997; Wu et al., 1997; see also Chapter 2 for more on caspases). Further, CED-4 that is complexed with CED-9 is unable to activate the processing of pro-CED-3, again emphasizing that CED-9 acts a moderator protein ‘upstream’ of CED-4 (Chaudhary et al., 1998; Chinnaiyan etal., 1997a). However, although CED-4 that is bound to CED-9 cannot initiate the processing of pro-CED-3, it can still bind to it because different domains in CED-4 are involved in the two different protein-protein interactions. The significance of this is discussed in Section 2.3.
Gain-of-function mutations in the egl-1 gene lead to the death of cells in the nervous system that normally survive whereas loss-of-function mutations cause the survival of all cells that normally die during hermaphrodite development (Trent et al., 1983; Desai et al., 1988; Conradt and Horvitz, 1998). However, egl-1 loss-of-function mutants are not like ced-3 and ced-4 loss-of-function mutants, despite the apparent similarity of the phenotypes. For example, worms carrying loss-of-function mutations in both ced-9 and in ced-3 or -4 exhibit inappropriate survival; worms carrying loss-of-function mutations in both ced-9 and egl-1 exhibit massive cell death: loss-of-function in egl-1 cannot suppress loss-of-function in ced-9 (Conradt and Horvitz, 1998). Thus, it is likely that egl-1 acts upstream of ced-9 (Figure 2). Finally it should be noted that the place of the ced-8 gene and the specific role of its protein product are not yet clear; the placing of ced-8 in the pathway in Figure 2 is an indication that it may be involved in some way at approximately that point.
2.2 Bodies of death: apoptosomes
In the previous section we have demonstrated that the proteins involved in bringing about developmentally regulated cell death in the somatic cells of C. elegans interact with each other in a very ordered manner. The question now is ‘how do these interacting proteins actually bring about apoptosis?’ One of us has recently developed a possible model that starts to answer this question (Hengartner, 1997, 1998a, 1998b, 1999). It is suggested, based on gene expression and protein distribution patterns, that CED-3, CED-4 and CED-9 are present in all cells whether or not they are destined to die. It is a key feature of the model that in most cells these proteins are associated together in a pre-assembled but inactive multiprotein ‘body of death’ or apoptosome. The apoptosome has been likened to a cocked gun which is safe unless it is disturbed in some way (Hengartner, 1999); it is safe because CED-9 is preventing CED-4 from initiating the activation of CED-3. So, what would make the complex ‘unsafe’? It is likely that CED-4 must be released from its binding by CED-9. The EGL-1 protein is a good candidate for being...
Table of contents
- Cover
- Halftitle Page
- Series Page
- Title Page
- Copyright Page
- Contents
- Contributors
- Abbreviations
- Preface
- 1 Apoptotic cell death: from worms to wombats … but what about the weeds? — M.O. Hengartner and J.A. Bryant
- 2 Caspases — at the cutting edge of cell death — H.R. Stennicke
- 3 Modulation of death receptor signalling — P. Schneider and J. Tschopp
- 4 The role of the Bcl-2 family in the modulation of apoptosis — C. Pepper and P. Bentley
- 5 The role of granzymes and serpins in regulating cell growth and death — P. Coughlin, E. Morris and L. Hampson
- 6 Mitochondria and cell death: a pore way to die? — A.P. Halestrap, J.P. Gillespie, A. O’Toole and E. Doran
- 7 Regulation of apoptosis by cell metabolism, cytochrome c and the cytoskeleton — J.M. Garland
- 8 Organelle-specific death pathways — K. X Pun and R.J. Brown
- 9 Telomeres, telomerase and cellular immortalization — C.J. Jones
- 10 Back from the brink: plant senescence and its reversibility — H. Thomas and I. Donnison
- 11 Senescence and cell death in Brassica napus and Arabidopsis — V. Buchanan-Wollaston and K. Morris
- 12 Cytokinin and its receptors — R. Hooley
- 13 Ethylene-triggered cell death during aerenchyma formation in roots — M.C. Drew, C.-J. He and P.W. Morgan
- 14 Targeted cell death in xylogenesis — M. C. McCann, N.J. Stacey and K. Roberts
- 15 Abscission and dehiscence — J.A. Roberts
- 16 Regulation of cell cycle arrest and cell death — alternative responses to DNA damage — C. Norbury
- 17 Tissue transglutaminase in cell death — M. Griffin and E. Verderio
- 18 Pathways used by adenovirus E1B 19K to inhibit apoptosis — K. Degenhardt, D. Perez and E. White
- 19 Tumour cell death — I.D. Bowen and F. Amin
- 20 Controlling apoptosis: implications for carcinogenesis — A.R. Clarke
- 21 Type I interferons inhibit the resolution of chronic inflammation — D. Scheel-Toellner, A.N. Akbar, D. Pilling, C.H. Orteu, CD. Buckley, K. Wang, P.R. Webb, J.M. Lord and M. Salmon
- 22 Death of plant cells and their contribution to rumen function — A.H. Kingston-Smith and M.K. Theodorou
- 23 Postscript: some concluding observations and speculations — S.G. Hughes and J.M. Garland
- Index