
Reviews in Computational Chemistry, Volume 32
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Reviews in Computational Chemistry, Volume 32
About this book
REVIEWS IN COMPUTATIONAL CHEMISTRY
THE LATEST VOLUME IN THE REVIEWS IN COMPUTATIONAL CHEMISTRY SERIES, THE INVALUABLE REFERENCE TO METHODS AND TECHNIQUES IN COMPUTATIONAL CHEMISTRY
Reviews in Computational Chemistry reference texts assist researchers in selecting and applying new computational chemistry methods to their own research. Bringing together writings from leading experts in various fields of computational chemistry, V olume 32 covers topics including global structure optimization, time-dependent density functional tight binding calculations, non-equilibrium self-assembly, cluster prediction, and molecular simulations of microphase formers and deep eutectic solvents. In keeping with previous books in the series, Volume 32 uses a non-mathematical style and tutorial-based approach that provides students and researchers with easy access to computational methods outside their area of expertise.
The chapters comprising Volume 32 are connected by two themes: methods that can be broadly applied to a variety of systems, and special considerations required when modeling specific system types. Each in-depth chapter contains background and theory, strategies for using the methods correctly, mini-tutorials and best practices, and critical literature reviews highlighting advanced applications. Essential reading for both newcomers and experts in the area of molecular modeling, this state-of-the-art resource:
- Covers topics such as non-deterministic global optimization (NDGO) approaches and excited-state dynamics calculations
- Contains a detailed overview of deep eutectic solvents (DESs) and simulation methods
- Presents methodologies for investigating chemical systems that form microphases with periodic morphologies such as lamellae and cylinders
- Features step-by-step tutorials on applying techniques to probe and understand the chemical dynamics exhibited in a system
- Includes detailed subject indices on each volume in the series and up-to-date compendiums of molecular modeling software, services, programs, suppliers, and other useful information
Reviews in Computational Chemistry, Volume 32 is a must-have guide for computational chemists, theoretical chemists, pharmaceutical chemists, biological chemists, chemical engineers, researchers in academia and industry, and graduate students involved in molecular modeling.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
1
NON‐DETERMINISTIC GLOBAL STRUCTURE OPTIMIZATION: AN INTRODUCTORY TUTORIAL
LIST OF ABBREVIATIONS
- ACO:
- Ant‐Colony Optimization
- BH:
- Basin Hopping (Monte Carlo with Minimization)
- CD:
- Collision Detection
- CD/DD:
- combined Collision and Dissociation Detection
- CSP:
- Crystal Structure Prediction
- DD:
- Dissociation Detection
- DFT:
- Density Functional Theory
- DFTB:
- Density Functional Tight‐Binding
- DOF:
- Degree of Freedom
- DGO:
- Deterministic Global Optimization
- FF:
- Force Field (empirical potential)
- GA:
- Genetic Algorithm
- GO:
- Global Optimization
- HPC:
- High‐Performance Computing
- IR:
- infrared
- :
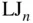
- Lennard‐Jones cluster with atoms
- MC:
- Monte Carlo
- MCM:
- Monte Carlo with Minimization (Basin Hopping)
- MD:
- Molecular Dynamics
- ML:
- Machine Learning
- MP2:
- Møller‐Plesset perturbation theory, second order
- MPI:
- message‐passing interface (parallelization)
- NDGO:
- Non‐Deterministic Global Optimization (algorithms)
- NFL(T):
- No Free Lunch (Theorem) for search and optimization
- PES:
- Potential Energy Surface
- PSO:
- Particle Swarm Optimization
- SA:
- Simulated Annealing
- wNFL(T):
- Weak No Free Lunch (Theorem)
INTRODUCTION
The Need for Structural Optimization
Search Space is Vast
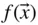
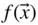
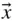
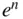
Table of contents
- Cover
- Table of Contents
- Title Page
- Copyright
- LIST OF CONTRIBUTORS
- PREFACE
- CONTRIBUTORS TO PREVIOUS VOLUMES
- 1 NON‐DETERMINISTIC GLOBAL STRUCTURE OPTIMIZATION: AN INTRODUCTORY TUTORIAL
- 2 DENSITY FUNCTIONAL TIGHT BINDING CALCULATIONS FOR PROBING ELECTRONIC‐EXCITED STATES OF LARGE SYSTEMS
- 3 ADVANCES IN THE MOLECULAR SIMULATION OF MICROPHASE FORMERS
- 4 MOLECULAR SIMULATIONS OF DEEP EUTECTIC SOLVENTS: A PERSPECTIVE ON STRUCTURE, DYNAMICS, AND PHYSICAL PROPERTIES
- INDEX
- Wiley End User License Agreement