![]()
CHAPTER 1
CLASSIFICATION OF MACROMOLECULES
1.1 COMPOSITION OF BIOMOLECULES
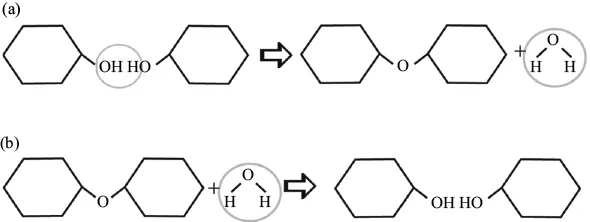
Macromolecules are the basic building blocks of living organisms. At a biochemical level, they can be defined as large organic molecules, or polymers, comprised of smaller molecules, known as monomers. Biological monomers have a carbon (C) backbone with varying degrees of oxygen (O), nitrogen (N), and hydrogen (H). Other elements are also present in organic biomolecules; however, the abundance and diversity varies throughout the cell and throughout species. Macromolecular polymers are formed as a result of dehydration synthesis reactions (Figure 1.1a), which form covalent bonds between monomers. Polymers can be broken down into monomers in the presence of water during hydrolysis reactions so that biosynthesis of new cellular molecules can proceed (Figure 1.1b).
Figure 1.1. Synthesis and hydrolysis of biological molecules. (a) Dehydration synthesis reactions result in the formation of covalent bonds between monomers to build larger macromolecules. (b) Hydrolysis reactions introduce a water molecule to break covalent bonds between polymers.
All biological macromolecules are carbon-based polymers, known as hydrocarbons. Each macromolecule differs based on the presence of varied side chains, or functional groups. The molecular composition of the functional group directly relates to the chemical properties of the molecule, and thus, the specific function of each macromolecule. Based on the complexity of the molecule and presence of functional groups, four distinct biological macromolecules have been categorized: carbohydrates, nucleic acids, proteins, and lipids. In this section, each macromolecule is described with respect to its molecular structure and cellular function.
1.2 NUCLEOTIDE STRUCTURE
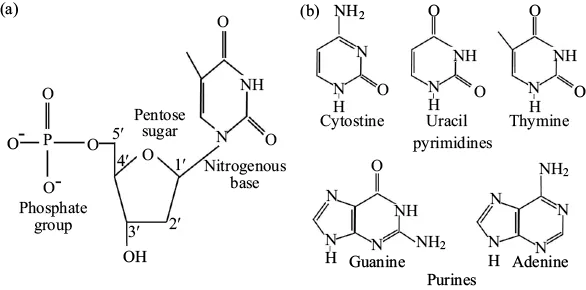
Nucleic acids are made of small monomers known as nucleotides (Figure 1.2a). Each nucleotide is comprised of a five-carbon sugar, known as a pentose. Each carbon of the pentose is numbered clockwise from oxygen with the corresponding number plus the (′) prime symbol. The numbering of carbons establishes molecular polarity and orientation of the other nucleotide subunits.
Figure 1.2. The structure of nucleotides (a) Nucleotides are carbon-based macromolecules that comprise nucleic acids. Each nucleotide is comprised of a pentose sugar attached to a nitrogenous base on the 1′ carbon and a phosphate group on the 5′ carbon. (c) There are five common nitrogenous bases found attached to the 1′ carbon of nucleotides. Pyrimidines (cytosine, thymine, and uracil) are single ringed; whereas, purines (adenine and guanine) are double-ringed.
The 1′ carbon is covalently bound to one of five nonpolar nitrogenous bases. Nitrogenous bases are also referred to as nitrogen-containing bases because they include a nitrogen atom that shares chemical properties with that of a base. Nitrogenous bases come in two flavors: double-ringed purines or single-ring pyrimidines (Figure 1.2b). Guanine (G) and adenine (A) are purines; whereas, cytosine (C), thymine (T), and uracil (U) are the most abundant pyrimidines found in nucleic acids.
Moving clockwise around the molecule, the 2′ and 3′ carbons are attached to either a hydrogen atom or a hydroxyl group (OH). The 5′ carbon is covalently bound to a phosphate group, which consists of a phosphorus bound to four oxygen atoms. The phosphate group attached to the 5′ carbon of the sugar on one nucleotide forms a covalent ester bond with the free hydroxyl on the 3′ carbon of the next nucleotide (Figure 1.2). These bonds are called phosphodiester bonds, and the positioning of nucleotides in this manner induces formation of a sugar-phosphate backbone.
1.2.1 MOLECULAR COMPOSITION OF DNA
Deoxyribonucleic acid (DNA) encodes the genetic instructions for the cell in just four letters—A,T,C, and G. These letters represent the nitrogenous bases found attached to nucleotides. Nucleotides that comprise DNA contain deoxyribose, a pentose sugar distinguished by a free hydroxyl at the 3′ position. DNA is double stranded, consisting of two linear sugar-phosphate backbones that run opposite each other and twist together into a helix. The two strands are antiparallel due to the opposing positions of the 5′ and 3′ carbons; therefore, the strands are designated as either 5′–3′ or 3′–5′ to distinguish one from the other. This gives the molecule polarity and plays a large factor in the replication process.
The sugar-phosphate backbone in DNA is negatively charged and hydrophilic, which promotes bonding with water. The helix is held together as a result of hydrogen bonding between the nitrogenous bases on opposing strands. In nearly every circumstance, adenine (A) will form two hydrogen bonds with thymine (T); whereas, guanine (G) will form three hydrogen bonds with cytosine (C). The relative amount of each nitrogenous base varies between different species; however, the bonding relationship is conserved from unicellular prokaryotic organisms up to complex multicellular organisms. The order and position of the nitrogenous bases corresponds to a molecular code, which serves as instructions for synthesizing all of the proteins and functional Ribonucleic acid (RNA) within a cell.
1.2.2 MOLECULAR COMPOSITION OF RNA
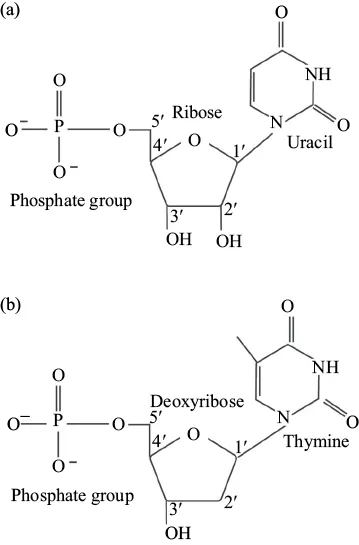
RNA is the single-stranded biochemical relative of DNA. Ribose is present in RNA, which can be distinguished from deoxyribose by the presence of hydroxyl groups on both the 2′ and the 3′ carbon (Figure 1.3a). Additional variation between DNA and RNA is found in the nitrogenous bases, as DNA possesses thymine (T), whereas RNA contains Uracil (U) (Figure 1.3b). The information stored in DNA is decoded by RNA, which is chemically similar, yet more diverse, in functionality. During gene expression, DNA is temporarily opened up by an enzyme known as RNA polymerase, which uses DNA sequences as a template for synthesizing a molecular copy in the form of RNA. RNA holds many roles and can be processed to relay different messages within the cell (more in Chapter 6). Recent research has highlighted certain RNAs as noncoding, meaning that they are never translated into protein and serve specific functions on their own such as catalysis and regulation of gene expression.
Figure 1.3. The structural variations between RNA and DNA. (a) RNA is comprised of ribonucleotides, each consisting of ribose sugar, a nitrogenous base (adenine, guanine, cytosine, or uracil) and a phosphate group. (b) DNA is comprised of deoxyribonucleotides, each of which contains deoxyribose, a nitrogenous base (adenine, guanine, cytosine, or thymine), and a phosphate group. Structurally both form phosphodiester bonds between the 3′ hydroxyl of one nucleotide and the 5′ carbon of another.
1.2.3 OTHER NUCLEIC ACIDS
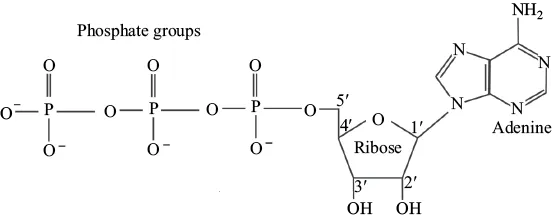
There are other nucleic acids that are essential to cell function and survival. These nucleic acids function primarily in energy storage and transfer, and serve as key players during cellular respiration. Perhaps the most noteworthy is adenosine triphosphate (ATP), which exists as a nucleotide monomer of ribose, adenine, and three phosphate groups (Figure 1.4). The instability of bonds between the phosphate groups allows rapid hydrolysis of the terminal phosphate to release energy and facilitate endergonic reactions.
Figure 1.4. The structure of adenosine triphosphate (ATP). ATP is a nucleoside triphosphate.
Nicotinamide adenine dinucleotide is another noteworthy nucleic acid. The substructure of this coenzyme is characterized as a dinucleotide due to the presence of only two nucleotides linked together via phosphodiester bonds. Nicotinamide adenine dinucleotide exists as either oxidized (NAD+) or reduced (NADH) and is essential for cellular respiration.
1.3 PROTEIN STRUCTURE AND FUNCTION
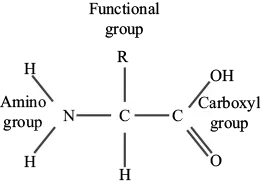
Proteins are the most diverse macromolecule with respect to shape, structure, and function. Comprised of amino acids, the varied nature of proteins is dictated by variation in functional group (R). There are 20 different amino acids; therefore, each is distinguished from one another by a distinct chemical side chain that dictates the bonding affinity and, thus, chemical behavior. In addition to the variable R group, all amino acids have a carboxyl group, an amino group, and a hydrogen atom attached to a central α carbon (Figure 1.5).
Figure 1.5. The structure of amino acids. Amino acids are carbon-based monomers of proteins. Each amino acid contains a central carbon bound to a hydrogen atom, a carboxyl group, an amino group, and a distinct functional (R) group. There are 20 different amino acids, each differentiated by the variable R group.
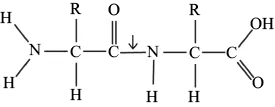
Dehydration synthesis reactions between the carboxyl and amino group of singular amino acids results in the formation of peptide bonds. Peptide bonds are incredibly strong and covalent in nature due to the sharing of valence electrons between C–N (Figure 1.6). Once peptide bonds are formed between amino acids, the sequences are characterized as residues. The sequence of amino acids determines the function of the final folded protein, as the interactions between the R groups contort the molecule into a distinct conformation.
Figure 1.6. Peptide bonds. Peptide bonds (arrow) form as a result of dehydration synthesis reactions between the amino groups and carboxyl groups of different amino acids.
1.3.1 PROTEIN FOLDING
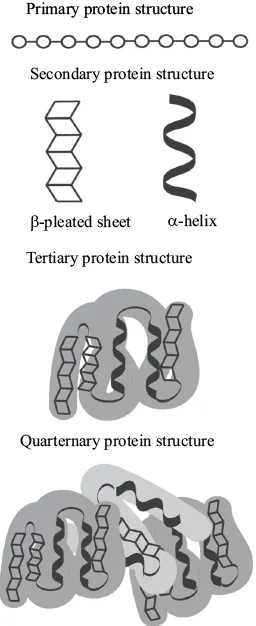
The unique sequence of amino acid residues represents the primary structure of protein folding (Figure 1.7). Given the 20 potential amino acids and unlimited possibilities for length and sequence, this structure is of particular importance because the organization of R groups drives bond formation during the remainder of protein folding. For example, the proximity of polar and nonpolar amino acids in the primary structure predicts how this protein will fold in later stages to protect hydrophobic residues from water and promote the interaction of hydrophilic residues with water.
Figure 1.7. Protein folding. The process of protein folding can be characterized by four distinct structures. The primary protein structure is characterized by the formation of peptide bonds between amino acids to form a polypeptide chain. The secondary structure is driven by hydrogen bonding between carboxyl and amino groups giving rise to the formation of either alpha-helices or beta pleated sheets. The tertiary structure forms as a result of bonding between R-groups giving rise to a 3-dimensional shape. The quaternary structure forms when two or more polypeptides bond to form a final functional protein.
The secondary structure of protein folding results from hydrogen bonding between the oxygen on the carboxyl group of one amino acid and th...