![]()
CHAPTER 1
Introduction and Overview
CONTENTS
1.1 The Biological Importance of DNA
1.2 The Origins of Nucleic Acids Research
1.3 Early Structural Studies on Nucleic Acids
1.4 The Discovery of the Structure of DNA
1.5 The Advent of Molecular Biology
1.6 The Partnership of Chemistry and Biology
1.7 Frontiers in Nucleic Acids Research
References
1.1 THE BIOLOGICAL IMPORTANCE OF DNA
From the beginning, the study of nucleic acids has drawn together, as though by a powerful unseen force, a galaxy of scientists of the highest ability.1,2 Striving to tease apart its secrets, these talented individuals have brought with them a broad range of skills from other disciplines while many of the problems they have encountered have proved to be soluble only by new inventions. Looking at their work, one is constantly made aware that scientists in this field appear to have enjoyed a greater sense of excitement in their work than is given to most. Why?
For over 60 years, such men and women have been fascinated and stimulated by their awareness that the study of nucleic acids is central to the knowledge of life. Let us start by looking at Fred Griffith, who was employed as a scientific civil servant in the British Ministry of Health investigating the nature of epidemics. In 1923, he was able to identify the difference between a virulent, S, and a non-virulent, R, form of the pneumonia bacterium. Griffith went on to show that this bacterium could be made to undergo a permanent, hereditable change from non-virulent to virulent type. This discovery was a bombshell in bacterial genetics.
Oswald Avery and his group at the Rockefeller Institute in New York set out to identify the molecular mechanism responsible for the change Griffith had discovered, now technically called bacterial transformation. They achieved a breakthrough in 1940 when they found that non-virulent R pneumococci could be transformed irreversibly into a virulent species by treatment with a pure sample of high molecular weight DNA.3 Avery had purified this DNA from heat-killed bacteria of a virulent strain and showed that it was active at a dilution of 1 part in 109.
Avery concluded that ‘DNA is responsible for the transforming activity’ and published that analysis in 1944, just 3 years after Griffith had died in a London air-raid. The staggering implications of Avery’s work turned a searchlight on the molecular nature of nucleic acids and it soon became evident that ideas on the chemistry of nucleic acid structure at that time were wholly inadequate to explain such a momentous discovery. As a result, a new wave of scientists directed their attention to DNA and discovered that large parts of the accepted tenets of nucleic acid chemistry had to be set aside before real progress was possible. We need to examine some of the earliest features of that chemistry to fully appreciate the significance of later progress.
1.2 THE ORIGINS OF NUCLEIC ACIDS RESEARCH
Friedrich Miescher started his research career in Tübingen by looking into the physiology of human lymph cells. In 1868, seeking a more readily available material, he began to study human pus cells, which he obtained in abundant supply from the bandages discarded from the local hospital. After defatting the cells with alcohol, he incubated them with a crude preparation of pepsin from pig stomach and so obtained a grey precipitate of pure cell nuclei. Treatment of this with alkali followed by acid gave Miescher a precipitate of a phosphorus-containing substance, which he named nuclein. He later found this material to be a common constituent of yeast, kidney, liver, testicular and nucleated red blood cells.4
After Miescher moved to Basel in 1872, he found the sperm of Rhine salmon to be a more plentiful source of nuclein. The pure nuclein was a strongly acidic substance, which existed in a salt-like combination with a nitrogenous base that Miescher crystallized and called protamine. In fact, his nuclein was really a nucleoprotein and it fell subsequently to Richard Altman in 1889 to obtain the first protein-free material, to which he gave the name nucleic acid.
Following William Perkin’s invention of mauveine in 1856, the development of aniline dyes had stimulated a systematic study of the colour-staining of biological specimens. Cell nuclei were characteristically stained by basic dyes, and around 1880, Walter Flemming applied that property in his study of the rod-like segments of chromatin (called so because of their colour-staining characteristic), which became visible within the cell nucleus only at certain stages of cell division. Flemming’s speculation that the chemical composition of these chromosomes was identical to that of Miescher’s nuclein was confirmed in 1900 by E.B. Wilson who wrote
Now chromatin is known to be closely similar to, if not identical with, a substance known as nuclein which analysis shows to be a tolerably definite chemical compound of nucleic acid and albumin. And thus we reach the remarkable conclusion that inheritance may, perhaps, be affected by the physical transmission of a particular compound from parent to offspring.
While this insight was later to be realized in Griffith’s 1928 experiments, all of this work was really far ahead of its time. We have to recognize that, at the turn of the century, tests for the purity and identity of substances were relatively primitive. Emil Fischer’s classic studies on the chemistry of high molecular weight, polymeric organic molecules were in question until well into the twentieth century. Even in 1920, it was possible to argue that there were only two species of nucleic acids in nature: animal cells were believed to provide thymus nucleic acid (DNA), while nuclei of plant cells were thought to give pentose nucleic acid (RNA).
1.3 EARLY STRUCTURAL STUDIES ON NUCLEIC ACIDS
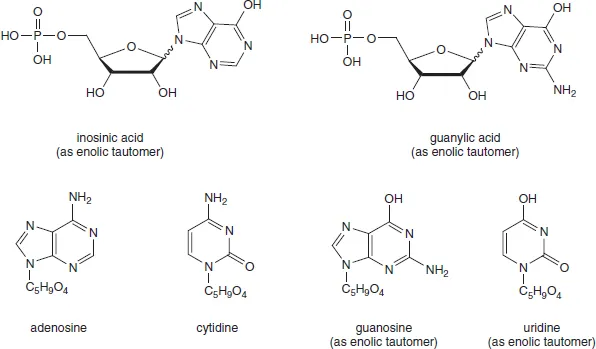
Accurate molecular studies on nucleic acids essentially date back to 1909 when Levene and Jacobs began a reinvestigation of the structure of nucleotides at the Rockefeller Institute. Inosinic acid, which Liebig had isolated from beef muscle in 1847, proved to be hypoxanthine-riboside 5′-phosphate. Guanylic acid, isolated from the nucleoprotein of pancreas glands, was identified as guanine-riboside 5′-phosphate (Figure 1.1). Each of these nucleotides was cleaved by alkaline hydrolysis to give phosphate and the corresponding nucleosides, inosine and guanosine, respectively. Since then, all nucleosides are characterized as the condensation products of a pentose and a nitrogenous base while nucleotides are the phosphate esters of one of the hydroxyl groups of the pentose.
Figure 1.1 Early nucleosides and nucleotide structures (using the enolic tautomers originally employed). Wavy lines denote unknown stereochemistry at C-1′
Thymus nucleic acid, which was readily available from calf tissue, was found to be resistant to alkaline hydrolysis. It was only successfully degraded into deoxynucleosides in 1929 when Levene adopted enzymes to hydrolyse the deoxyribonucleic acid followed by mild acidic hydrolysis of the deoxynucleotides. He identified its pentose as the hitherto unknown 2-deoxy-D-ribose. These deoxynucleosides involved the four heterocyclic bases, adenine, cytosine, guanine and thymine, with the latter corresponding to uracil in ribonucleic acid.
Up to 1940, most groups of workers were convinced that hydrolysis of nucleic acids gave the appropriate four bases in equal relative proportions. This erroneous conclusion probably resulted from the use of impure nucleic acid or from the use of analytical methods of inadequate accuracy and reliability. It led, naturally enough, to the general acceptance of a tetranucleotide hypothesis for the structure of both thymus and yeast nucleic acids, which materially retarded further progress on the molecular structure of nucleic acids.
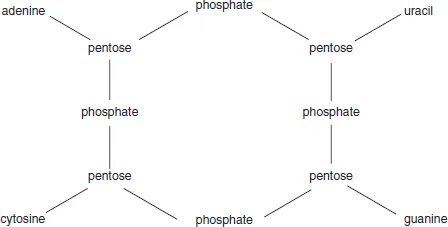
Several of these tetranucleotide structures were proposed. They all had four nucleosides (one for each of the bases) with an arbitrary location of the two purines and two pyrimidines. They were joined together by four phosphate residues in a variety of ways, among which there was a strong preference for phosphodiester linkages. In 1932, Takahashi showed that yeast nucleic acid contained neither pyrophosphate nor phosphomonoester functions and so disposed of earlier proposals in preference for a neat, cyclic structure which joined the pentoses exclusively using phosphodiester units (Figure 1.2). It was generally accepted that these bonded 5′- to 3′-positions of adjacent deoxyribonucleosides, but the linkage positions in ribonucleic acid were not known.
Figure 1.2 The tetranucleotide structure proposed for nucleic acids by Takahashi (1932)
One property stuck out like a sore thumb from this picture: the molecular mass of nucleic acids was greatly in excess of that calculated for a tetranucleotide. The best DNA samples were produced by Einar Hammarsten in Stockholm and one of his students, Torjbörn Caspersson, who showed that this material was greater in size than protein molecules. Hammarsten’s DNA was examined by Rudolf Signer in Bern whose flow-birefringence studies revealed rod-like molecules with a molecular mass of 0.5–1.0 × 106 Da. The same material provided Astbury in Leeds with X-ray fibre diffraction measurements that supported Signer’s conclusion. Finally, Levene estimated the molecular mass of native DNA to be between 200,000 and 1 × 106 Da, based on ultracentrifugation studies.
The scientists compromised. In his Tilden Lecture of 1943, Masson Gulland suggested that the concept of nucleic acid structures of polymerized, uniform tetranucleotides was limited, but he allowed that they could ‘form a practical working hypothesis’.
This then was the position in 1944 when Avery published his great work on the transforming activity of bacterial DNA. One can sympathize with Avery’s hesitance to press home his case. Levene, in the same Institute, and others were strongly persuaded that the tetranucleotide hypothesis imposed an invariance on the structure of nucleic acids, which denied them any role in biological diversity. In contrast, Avery’s work showed that DNA was responsible for completely transforming the behaviour of bacteria. It demanded a fresh look at the structure of nucleic acids.
1.4 THE DISCOVERY OF THE STRUCTURE OF DNA
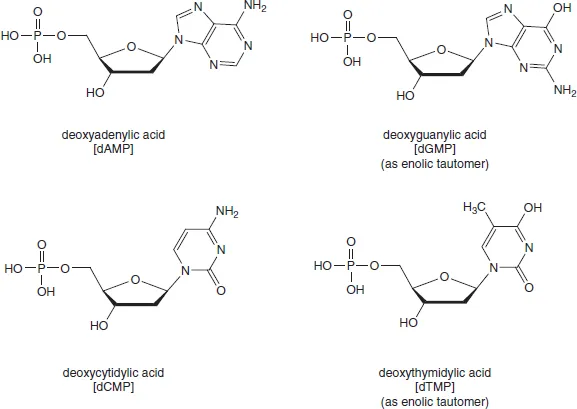
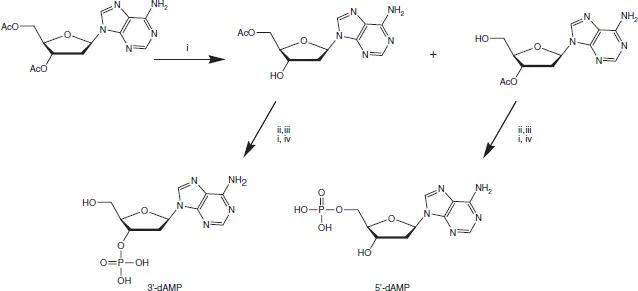
From the outset, it was evident that DNA exhibited greater resistance to selective chemical hydrolysis than did RNA. So, the discovery in 1935 that DNA could be cut into mononucleotides by an enzyme doped with arsenate was invaluable. Using this procedure, Klein and Thannhauser obtained the four crystalline deoxyribonucleotides, whose structures (Figure 1.3) were later put beyond doubt by total chemical synthesis by Alexander Todd5 and the Cambridge school he founded in 1944. Todd established the D-configuration and the glycosylic linkage for ribonucleosides in 1951, but found the chemical synthesis of the 2′-deoxyribonucleosides more taxing. The key to success for the Cambridge group was the development of methods of phosphorylation, for example for the preparation of the 3′- and 5′-phosphates of deoxyadenosine6 (Figure 1.4).
Figure 1.3 Structures of 5′-deoxyribonucleotides (original tautomers for dGMP and dTMP)
Figure 1.4 Todd’s synthesis of deoxyadenosine 3′- and 5′-phosphates Reagents: (i) MeOH, NH3 (ii) (PhO)2P(O) OP(H)(O)OCH2Ph (iii) N-chlorosuccinimide (iv) H2/PdC. (D.H. Hayes, A.M. Michelson and A.R. Todd, J. Chem. Soc., 1955, 808–815.)
All the facts were now available to establish the primary structure of DNA as a linear polynucleotide in which each deoxyribonucleoside is linked to the next by means of a 3′- to 5′-phosphate diester (see Figure 2.15). The presence of only diester linkages was essential to explain the stability of DNA to chemical hydrolysis, since phosphate triesters and monoesters, not to mention pyrophosphates, are more labile. The measured molecular masses for DNA of about 1 × 106 Da meant that a single strand of DNA would have some 3000 nucleotides. Such a size was much greater than that of enzyme molecules, but entirely compatible with Staudinger’s established ideas on macromolecular structure for synthetic and natural polymers. But by the mid-twentieth century, chemists could advance no further with the primary structure of DNA. Neither of the key requirements for sequence determination was to hand: there were no methods for obtaining pure samples of DNA with homogeneous base sequence nor were methods available for the cleavage of DNA strands at a specific base residue. Consequently, all attention came to focus on the sec...