![]()
Part I
Principles of Microbial Biotechnology
The use of microorganisms for large-scale industrial purposes has a long history, which is long before the realization of the activities of the microorganisms. For centuries, beer, wine, vinegar, soy sauce and other fermented foods were produced through spontaneous fermentation of natural occurring microorganisms or the use of carry-over microbial seeds from the previous batch of production. The quality and productivity of these early products were very often inconsistent. The development of scientific screening and isolation methods allows the selection of desirable natural occurring or mutated microorganisms for specific purposes. These methods, coupled with the advancement of the technical know-how in large-scale sterilization of culture media, in provision of adequate oxygen supply and in mixing homogeneity of the culture systems, enable the exploitation of both anaerobic (yeast and some bacteria) and aerobic microorganisms (fungi and some bacteria). Common examples are: the development of large-scale processes for the production of citric acid, amino acids and antibiotics; in improved biotransformation of steroid hormones, as well as the mass production of many enzymes. The diverse catalytic activities of microorganisms are being used more and more widely to perform specific chemical reactions in the industrial production processes.
Microbial biotechnology was pushed to a new height in the eighties when continuous fermentation and airlift fermentation processes were developed for the production of food and feed grade microbial protein from industrial by-products, such as methanol and alkanes. These processes lead to considerable savings in capital, energy and labor costs.
Modern techniques of gene manipulation, and advanced bioinformatics and biocomputing are powerful tools for genomic, proteomic and metabo-lomic research. The scientific breakthroughs that ensued have made feasible the industrial manufacturing of non-microbial products, such as human growth hormone, interferon and viral vaccines; and defined approaches in the monitoring and control of fementation processes.
It is the aim of Part I of this book to provide the readers with in-depth and comprehensive scientific knowledge in the areas as listed below, so as to facilitate the understanding of the various applications of micro-organisms and the production of their bioactive molecules in the biotechnological systems:
• Screening for microbial products
• Bioprocess technology
• Enzymology
• Manipulation of genes and metabolic pathways
• Application of bioinformatics and biocomputing.
![]()
Chapter 1
Microbial Screening
Allan Lim
Nestlé R&D Center Pte Ltd
15A, Changi Business Park Central 1
#05-02/03 Eightrium@Changi Business Park, Singapore 486035
Tan Hai-Meng
Kemin Industries (Asia) Pte Ltd
12 Senoko Drive, Singapore 758200
1.1. Introduction
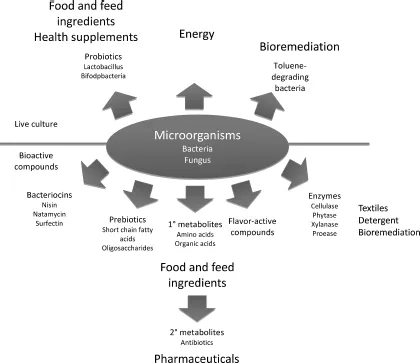
Microbes can be harnessed as “mini-factories” to produce primary and secondary metabolites for human applications such as ethanol for breweries, citric acid for food, and antibiotics for disease prophylaxis and treatment (Fig. 1.1). Since most of the metabolic pathways in microorganisms are very similar, the primary objective of screening is to search for the strain(s) that is most efficient and cost effective in producing these metabolites. Other considerations such as safety and sensory profiles are particularly important in the food and beverage industries. For instance, enormous resources are used to isolate special yeast strains (e.g. Saccharomyces uvarum (carlsbergenesis) and Torulaspora delbrueckii) that confer unique flavors to beers. In contrast, microbes for non-food applications have much less restrictions, and they can even be mutants generated in laboratories. For example, genetically-modified Escherichia coli and Pseudomomas spp. are now used extensively in bioremediation. A screen is a systematic assay procedure that allows testing of numerous compounds for a particular activity, phenotype for a defined
Fig. 1.1. Application of microorganisms in various industries.
application. The better the design of the screen, the higher the chance of finding the strain in the shortest time. Access to unique or previously unculturable microorganisms will also ensure the success of screening. Many companies even patent sources or screening procedures for competitive advantage, such as the process of “bioprospecting” a proprietary genetic bank of microbes from extreme ecosystems (e.g. volcanoes and deep sea hydrothermal vents). Proprietary screening technology will then facilitate rapid discovery and evolution of unique and tailored enzymes from this genetic bank to suit various challenging industrial environments. Companies are also known to partner with governmental research institutions (e.g. BIOTEC, National Centre for Genetic Engineering and Biotechnology, Thailand) to screen thousands of enzymes and microorganisms from diverse environments for possible applications in disease treatment and drug development. It is usual for such collaborations to have the country compensated either in monetary or non-monetary means,
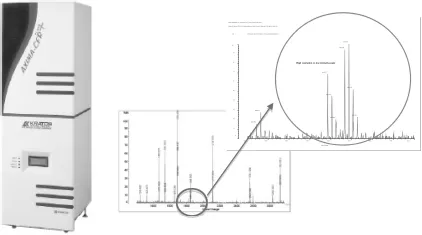
Fig. 1.2. High Performance MALDI-TOF MS designed for the mass analysis of biomol-ecules. High quality peptide mapping fingerprints (PMF) could be generated easily and resolved at high resolution. (Courtesy of Shimadzu (Asia Pacific) Pte Ltd.)
such as capacity building and technology transfer. As legal owners of these microorganisms and natural compounds, the research institutions will also continue to evaluate them in their home country for applications outside the scope of the commercial collaboration.
The scale and speed of screening has also been dramatically increasing at an unprecedented speed due to the availability of advanced instrumentations, such as the MALDI-TOF/TOF and LC-MALDI for high throughput screening of metabolites (Fig. 1.2).
Although high throughput screening was primarily developed for drug screening, it is increasingly being deployed in screening of microbials for the target metabolites. Today, this is often coupled with high throughput DNA sequencing so that shortlisted microbial strains can be rapidly characterized.
1.2. Screening Process
Several key areas are involved in the screening process of microorganisms and their metabolites. The first is the development and validation of assay(s), which upon further assessment for its reproducibility and compatibility with the physicochemical characteristics of the samples converting it to a primary or pilot screen. The primary screening process makes possible the estimation of hit rates and for large numbers of samples to be screened rapidly to obtain sufficient data to initiate a full screening program. The second area is high throughput screening, which enables large numbers of potential samples to be screened with minimum numbers of steps using semi- or fully automated systems. The high throughput screening system will also perform retesting and confirmation of active metabolites from the initial screen. The third area is secondary screening of the active metabolites or “hits” from the primary screen to eliminate false positive results. At this stage, active fractions are further characterized based on heat stability, chemical classes, molecular weights, pH optimum and enzyme inhibition patterns. Following purification and structure elucidation of active metabolites, the potential molecules are then subjected to further in vivo testing.
1.3. Screening Methods
The use of appropriate isolation procedures is an important initial step in the development of preliminary screens for useful secondary metabolites. Basically, this involves the selective isolation of producer-bacteria and a demonstration, usually visual, of their secondary metabolites. Following selection and pre-treatment of the material containing bacterial strains of interest, specific selective laboratory media are used for the growth and isolation of the microorganisms of interest. Pre-treatment procedures used to increase the relative number of producer microorganisms e.g. antibiotic-producing actinomycetes relative to bacteria on isolation plates can include processes of simple drying and dry heating to reduce the number of undesired Gram-negative bacteria. The latter can cause swarming on the isolation plates but the aerial spores of most actinoycetes are relatively resistant to desiccation. Incubation conditions generally include temperatures of 25–30°C and periods of 7–14 days although longer incubation periods may be needed for slower growing strains. In some cases, the incorporation of spore activating and bactericidal agents in isolation media such as sodium doedcyl sulphate, phenol, antibiotics and yeast extract in combination with dry heat can help increase the recovery of actinomycetes while suppressing the counts of bacteria. Antibiotics such as rifampicin, novobiocin and oxytetracylcine are commonly used for the isolation of Acitnomadura, Micromonospora and Streptoverticillium, respectively. At the same time, antifungal agents such as cycloheximide and nyastatin which do not inhibit actinomycetes are regularly added to eliminate fungal contamination. To help in the isolation of specific taxa of actinomycetes, chromogenic or fluorogenic substrates for genus specific enzymes can be incorporated into the selective media used.
Once the microorganisms of interest have been isolated, several methods can be used to investigate the antimicrobial activities of secondary metabolites produced by these microorganisms. This can involve overlaying the isolation plate with a single test or indicator organism. Another variation, also known as the agar-spot test, has also been used for the detection of antagonistic activity between bacteria. These methods can be quite laborious since only one indicator organism at a time can be applied to each isolation plate. As an alternative, replica plating of selected colonies can be used to study a range of indicator organisms. However, the spread of motile bacteria on the surface of each isolation plate can limit the usefulness of this method.
The sensitivity of an isolation medium can be influenced by its nutrient composition, pH, selective agents and incubation temperature. Typically, nonselective and/or selective enrichments are required to increase the sensitivity of detection of target bacteria. Generally, the stepwise enrichment process begins with pre-enrichment or primary enrichment, in which contaminated samples are incubated in a nutritious nonselective medium to allow for the growth of target bacteria. Pre-enriched samples are then transferred to a secondary selective enrichment medium, where the normal flora is suppressed but the target bacteria is allowed to grow. Finally, the producer bacteria is streaked on selective isolation agar and isolated as a pure culture. Pure producer bacterial culture is then transferred into a broth system to begin the fermentation and optimization process of enhancing the production of active secondary metabolites. Filtration of fermented broth through a membrane filter (0.20–0.45 μm pore size) is routinely used to separate cells from the aqueous phase. The filtrate is then subjected to screening against a spectrum of indicator organisms. The disk-activity assay has been the method of choice used in the comparison of relative activities of active filtrates from various bacterial fermented broths. Sterile analytical paper disks (0.5 inch in diameter) are immersed in sample filtrate and placed onto the surface of solidified agar medium seeded with an indicator organism. Following overnight incubation at a specific temperature, the relative activities between different filtrates can be observed from size of the zone of inhibition on each medium seeded with indicator organisms. Alternatively, a modified agar-well diffusion method can be used to examine the activity of the active filtrates from various bacterial fermented broths. In this method, wells are aseptically created by a hole-borer in a solidified medium seeded with indicator organisms. Similarly, the relative activities between different filtrates are determined based on the sizes of the zone of inhibition on the solidified medium. However, both techniques using the disk and well assays are laborious and time-consuming when screening the active filtrate against a wide range of bacteria. Therefore, to enable rapid screening of active samples, screens may be carried out using semi- or fully automated high ...