
Frontiers In Autism Research: New Horizons For Diagnosis And Treatment
New Horizons for Diagnosis and Treatment
- 724 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Frontiers In Autism Research: New Horizons For Diagnosis And Treatment
New Horizons for Diagnosis and Treatment
About this book
This book focuses on the emerging and expanding areas of research on ASD and their potential to lead to better diagnosis and more effective therapies. These areas include innovative and integrative approaches to genetic/genomic analyses and investigations of epigenetic contributions, including the role of noncoding RNAs, DNA methylation, alternative splicing, RNA editing, and faulty translation in gene regulation and expression, metabolic and immune dysfunction, co-morbidities, as well as hormonal and gene-environment interactions that may increase risk for ASD.
Within each chapter, experts review cutting-edge research as well as provide their perspective on the future of research in their respective areas, including the challenges involved and the types of studies or advances that are necessary to move the field forward to achieve predicted translational goals.
Contributors: Argel Aguilar-Valles, Evdokia Anagnostou, Emma Ashwin, Bonnie Auyeung, Kelly M Bakulski, Simon Baron-Cohen, Margaret L Bauman, Donna Betts, Chad A Bousman, Daniel B Campbell, Manuel F Casanova, Bhismadev Chakrabarti, Gursharan Chana, Abha Chauhan, Ved Chauhan, Jessica DeWitt, Keith W Dunaway, Alal Eran, Ian P Everall, M Daniele Fallin, Richard E Frye, Piers Gillett, Matthew Ginsberg, Christos G Gkogkas, Rhonda J Greenhaw, Simon G Gregory, Elena L Grigorenko, Feng Gu, Rebecca Harmer, Martha Herbert, Valerie W Hu, Karen L Jones, Petra Kern, Arkady Khoutorsky, Rebecca Knickmeyer, Isaac S Kohane, Louis M Kunkel, Janine M LaSalle, Michael V Lombardo, Deepali Mankad, Marvin Natowicz, Laura Nicholls, Christos Pantelis, Natalia Rakhlin, Radhika Ramadas, Daniel A Rossignol, Tewarit Sarachana, Stephen W Scherer, Gabriela Schmulevich, Ayten Shah, Frank R Sharp, Alison B Singer, Efstratios Skafidas, Estate M Sokhadze, Nahum Sonenberg, Boryana Stamova, Zohreh Talebizadeh, Renee Testa, Judy Van de Water, Irina Voineagu, Daniel Williams, Ryan K C Yuen, Daniela Zantomio.
Contents:
- Dissecting the Genetic Architecture of ASD:
- Phenotype Definition: A Cornerstone of Autism Research, Diagnosis and Therapy (Valerie W Hu)
- From Molecular Pathways to ASD Therapy: Insights from Syndromic Forms of Autism (Laura Nicholls, Radhika Ramadas and Irina Voineagu)
- Language Impairment in Autism Spectrum Disorders (Natalia Rakhlin and Elena L Grigorenko)
- Whole Genome Sequencing in Autism: Clinical Translation (Ryan K C Yuen and Stephen W Scherer)
- The Impact of Integrative Unconventional Data Analysis Approaches on Advancing Autism Genetics Research (Zohreh Talebizadeh and Ayten Shah)
- Construction of a Genetic Classifier for ASD Using Gene Pathway Analysis (Gursharan Chana, Renee Testa, Piers Gillett, Daniel Williams, Chad A Bousman, Daniela Zantomio, Ian P Everall, Christos Pantelis and Efstratios Skafidas)
- Gene Dysregulation in ASD: From Transcription to Translation:
- Genome-Wide Expression Studies of Blood and Lymphoblastoid Cell Lines in Autism Spectrum Disorders (Boryana Stamova and Frank R Sharp)
- Searching in the “Dark” Non-Coding RNA as a New Avenue of Autism Research (Tewarit Sarachana and Valerie W Hu)
- Targeting Noncoding RNA for Treatment of Autism Spectrum Disorders (Jessica DeWitt and Daniel B Campbell)
- A-to-I RNA Editing in Autism Spectrum Disorder (Alal Eran, Isaac S Kohane and Louis M Kunkel)
- Translational Control of Autism and Fragile-X Syndrome (Christos G Gkogkas, Argel Aguilar-Valles, Arkady Khoutorsky and Nahum Sonenberg)
- Epigenetic, Environmental, and Physiological Contributions to ASD:
- Epigenetics in Autism (Matthew Ginsberg and Marvin Natowicz)
- The Epigenetics of Autism — Running Beyond the Bases (Simon G Gregory)
- Genes and Environment in Autism Spectrum Disorders: An Integrated Perspective (Kelly M Bakulski, Alison B Singer and M Daniele Fallin)
- The Potential Brain Drain from Environmental Exposures on the Methylome and Genome Across Generations (Janine M LaSalle and Keith W Dunaway)
- Oxidative Stress and Mitochondrial Dysfunction in ASDs (Feng Gu, Ved Chauhan and Abha Chauhan)
- Maternal Autoantibodies in Autism Spectrum Disorder (Karen L Jones and Judy Van de Water)
- Why is Autism More Common in Males? (Simon Baron-Cohen, Michael V Lombardo, Bonnie Auyeung, Emma Ashwin, Bhismadev Chakrabarti and Rebecca Knickmeyer)
- Moving Towards Personalized Treatment of ASD and Lifespan Issues:
- Future Directions in Psychopharmacology of Autism Spectrum Disorder (Deepali Mankad and Evdokia Anagnostou)
- Medical Co-Morbidities in Autism: Clues to Underlying Biological Mechanisms and/or Diagnostic Subtypes? (Margaret L Bauman)
- Translational Implications of a Whole-Body Approach to Brain Health in Autism: How Transduction Between Metabolism and Electrophysiology Points to Mechanisms for Neuroplasticity (Martha R Herbert)
- Achieving Optimal Outcomes in Autism: Treating Potentially Reversible Conditions Associated with Autism Spectrum Disorder (Richard E Frye and Daniel A Rossignol)
- Transcranial Magnetic Stimulation: Application in Autism Treatment (Manuel F Casanova and Estate M Sokhadze)
- Music Therapy: Personalized Interventions for Individuals with Autism Spectrum Disorder (Petra Kern)
- The Contributions of Art Therapy in Treatment, Assessment, and Research with People Who have Autism Spectrum Disorders (Donna Betts, Rebecca Harmer and Gabriela Schmulevich)
- Shifting Paradigms: An Examination of Our Understanding of Adult Autism (Rhonda J Greenhaw)
Readership: Established investigators and students engaged in autism research; also the general population, especially families of individuals affected by ASD and their professional caregivers.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
Chapter 12
Epigenetics in Autism
Introduction
Fundamentals of Epigenetics
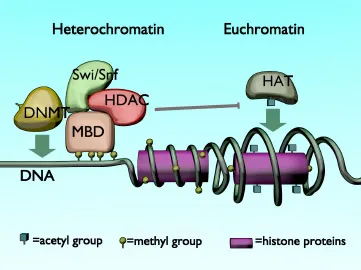
Table of contents
- Cover Page
- Title Page
- Copyright Page
- Dedication
- Contents
- Preface
- About the Editor
- List of Contributors
- Dissecting the Genetic Architecture of ASD
- Gene Dysregulation in ASD: From Transcription to Translation
- Epigenetic, Environmental, and Physiological Contributions to ASD
- Moving Towards Personalized Treatment of ASD and Lifespan Issues
- Index