
- 480 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Orchid Biotechnology Iii
About this book
-->
This book provides a first hand and complete information on orchid biotechnology for orchid lovers, graduate students, researchers and industry growers. It contains comprehensive genomics and transcriptomics data, and a thorough discussion of the molecular mechanism of orchid floral morphogenesis. The contributors to the book are all orchid enthusiasts with more than 20 years' experience in the field.
With more than 25,000 species, orchids are the most species-rich of all angiosperm families. They show wide diversity of epiphytic and terrestrial growth forms and have successfully colonized almost every habitat on earth. Orchids are fantastic for their spectacular flowers with highly evolved petal, labellum, and fused androecium and gynoecium, gynostemium, to attract pollinators for effective pollination. In addition, orchids have attracted the interest of many evolutionary biologists due to their highly specialized evolution and adaptation strategies.
Orchid Biotechnology III covers the most update knowledge of orchid biotechnology research on Phalaenopsis, Oncidium, Cymbidium, Anoectohilus, Paphiopedilum, and Erycina pusilla. It will provide graduate students, researchers, orchid lovers and breeders with an opportunity to understand the mechanism why the orchids are so mysterious and spectacular. Hopefully, this information will be helpful for breeders to enhance orchid breeding and create even more elegant and grace flowers.
-->
Contents: Genome Size Variation in Species of the Genus Phalaenopsis Blume (Orchidaceae) and Its Application in Variety Improvement (Wen-Huei Chen and Ching-Yan Tang);Distinct Distribution Patterns of 45S rDNA and 5S rDNA-NTS–Related Repeats Display Diverse Karyotypes in Paphiopedilum (Yung-I Lee, Fang-Chi Chang, and Mei-Chu Chung);Chromosome Number Variation in the Emergent Orchid Model Species Erycina pusilla (Hsuan-Yu Yeh, Choun-Sea Lin, and Song-Bin Chang);Comparative Chloroplast DNA Analysis of Phalaenopsis Orchids and Evaluation of cpDNA Markers for Distinguishing Moth Orchids (Ching-Chun Chang, Wen-Luan Wu, Yu-Chang Liu, Cheng-Fong Jheng, Tien-Chih Chen, Bo-Yen Lin, Jhong-Yi Lin, Ting-Chieh Chen, and Yuech-Feng Lee);Development of SSR Markers in Phalaenopsis Orchids, Their Characterization, Cross-Transferability and Application for Identification (Yu-Lin Chung, Yi-Tzu Kuo, and Wen-Luan Wu);Warm-Night Temperature Delays Spike Emergence and Alters Carbon Pool Metabolism in the Stem and Leaves of Phalaenopsis aphrodite (Yo-Ching Liu, Kai-Meng Tseng, Ching-Chia Chen, Yun-Tai Tsai, Cheng-Huan Liu, Wen-Huei Chen, and Heng-Long Wang);Light-Emitting Diodes Affect the Growth and Gene Expression of Phalaenopsis Plantlets In Vitro (Huey-Wen Chuang);Handling, Packaging, and Transportation of Phalaenopsis Plants for Marine Shipment (Chao-Chia Huang, Ching-Chieh Huang, and Pen-Chih Lai);The Orchid-Infecting Viruses Found in the 21st Century (Chia-Hwa Lee, You-Xiu Zheng and Fuh-Jyh Jan); Phalaenopsis Viruses and Their Detection Techniques (Chin-An Chang);Virus Resistance in Orchids (Kah Wee Koh and Ming-Tsair Chan);Perianth Regulation in Orchids (Hsing-Fun Hsu and Chang-Hsien Yang);Flower Development of Phalaenopsis Orchids Involves Functionally Divergent B-Class and E-Class MADS-Box Genes (Zhao-Jun Pan, Wen-Chieh Tsai, and Hong-Hwa Chen);The Function of C/D-Class MADS Box Genes in Orchid Gynostemium and Ovule Development (You-Yi Chen and Wen-Chieh Tsai);The Dual Function of Harpin in Disease Resistance and Growth in Phalaenopsis Orchids (Huey-Wen Chuang);Development of a High-Throughput Virus-Induced Gene-Silencing Vector in Phenotypic Screening of Orchids (Ho-Hsiung Chang, Li Chang, Hsiang-Chia Lu, and Hsin-Hung Yeh);Application of VIGS to Floral Gene Function Studies of the Orchid, a Nonmodel Plant (Ming-Hsien Hsieh and Hong-Hwa Chen);Functional Study of Phytoene Synthase by RNAi-Based Downregulation in the Oncidesa Orchid (Chao-Wei Yeh, Jianxin Liu, Chung-Yi Chiou, Chin-Hui Shen, Peng-Jen Chen, Jung-Chung Liou, Chin-Der Jian, Xiao-Lan Shen, Fu-Quan Shen, and Kai-Wun Yeh);Flower Color and Pigmentation Patterns in Phalaenopsis Orchids (Chia-Chi Hsu and Hong-Hwa Chen);Prolonged Exposure to Elevated Temperature Induces Floral Transition via Upregulation of Cytosolic Ascorbate Peroxidase 1 and Subsequent Reduction of the Ascorbate Redox Ratio in the Oncidium Hybrid Orchid (Dan-Chu Chin and Kai-Wun Yeh);Methylation Effect on Chalcone Synthase Gene Expression Determines Anthocyanin Pigmentation in Floral Tissues of Two Oncidium Orchid Cultivars (Dan-Chu Chin and Kai-Wun Yeh); --> -->
Readership: Graduate students and researchers in agricultural sciences (crop sciences, plant biology, biotechnology, genetics & genomics, microbiology & virology.
-->Orchid, Biotechnology, Flower, Genomics, Virus-Induced Gene Silencing, Morphogenesis, Resistance, Transformation, Virus, Database, Chloroplast, SSR Marker, MADS-Box Genes, Ambient Temperature, LED Light, Chromosome, Pigmentation Patterning, Promoters, Redox
- This book provides the first hand and complete information of orchid biotechnology for orchid lovers, graduate students, researchers and industry growers
- It contains comprehensive genomics and transcriptomics data, and thorough discussion of molecular mechanism of orchid floral morphogenesis
- The contributors for chapters are all endeavor in orchid research for more than 20 years
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
CHAPTER 1
Genome Size Variation in Species of the Genus Phalaenopsis Blume (Orchidaceae) and Its Application in Variety Improvement*
Wen-Huei Chen† and Ching-Yan Tang
1.1.Introduction
1.2.Variation of the Nuclear DNA Content in Phalaenospsis Species Using the PI Fluorochrome
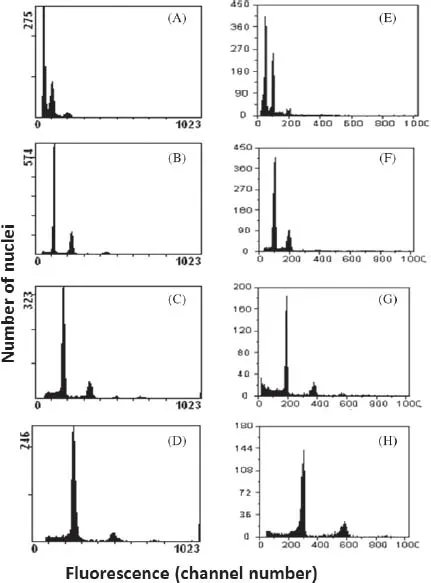

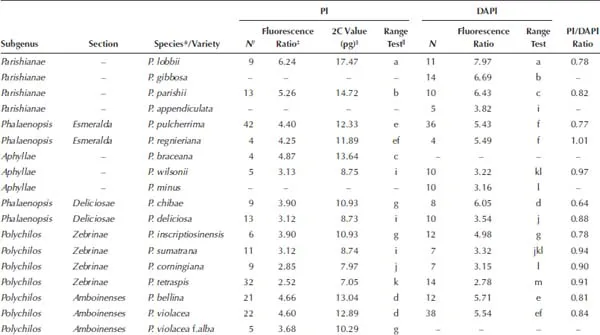
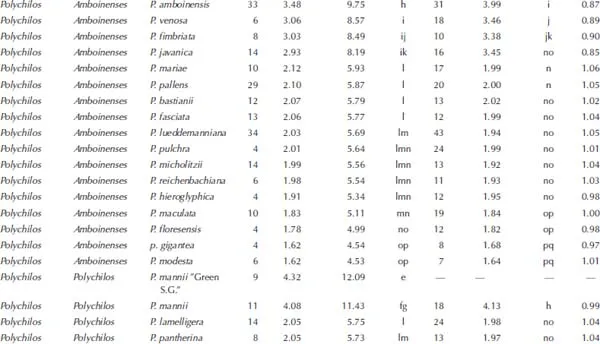
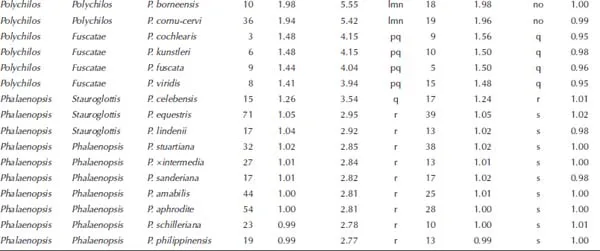
†Number of measurements.
‡Fluorescence ratio = sample G1 peak mean / standard (diploid P. aphrodite) G1 peak mean.
§2C values of 50 species measured by PI were calibrated by using diploid P. aphrodite (2C value = 2.80 pg) as the standard.
||The Mean value for a particular species followed by the same letters within a column is not significantly different at α = 0.05 (Tukey–Kramer′s test).
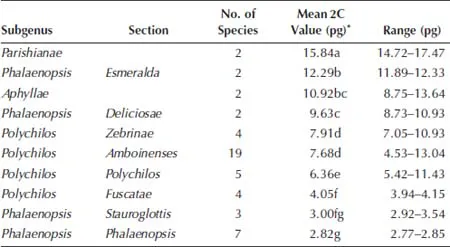
1.3.Comparison of the Variations of the Fluorescence Ratios in Phalaenopsis Species Using the DAPI and PI Fluorochromes
Table of contents
- Cover
- Halftitle
- Title
- Copyright
- Foreword
- Preface
- Contents
- Chapter 1 Genome Size Variation in Species of the Genus Phalaenopsis Blume (Orchidaceae) and Its Application in Variety Improvement
- Chapter 2 Distinct Distribution Patterns of 45S rDNA and 5S rDNA-NTS–Related Repeats Display Diverse Karyotypes in Paphiopedilum
- Chapter 3 Chromosome Number Variation in the Emergent Orchid Model Species Erycina pusilla
- Chapter 4 Comparative Chloroplast DNA Analysis of Phalaenopsis Orchids and Evaluation of cpDNA Markers for Distinguishing Moth Orchids 61 Ching-Chun Chang, Wen-Luan Wu, Yu-Chang Liu,
- Chapter 5 Development of SSR Markers in Phalaenopsis Orchids, Their Characterization, Cross-Transferability and Application for Identification
- Chapter 6 Warm-Night Temperature Delays Spike Emergence and Alters Carbon Pool Metabolism in the Stem and Leaves of Phalaenopsis aphrodite
- Chapter 7 Light-Emitting Diodes Affect the Growth and Gene Expression of Phalaenopsis Plantlets In Vitro
- Chapter 8 Handling, Packaging, and Transportation of Phalaenopsis Plants for Marine Shipment
- Chapter 9 The Orchid-Infecting Viruses Found in the 21st Century
- Chapter 10 Phalaenopsis Viruses and Their Detection Techniques
- Chapter 11 Virus Resistance in Orchids
- Chapter 12 Perianth Regulation in Orchids
- Chapter 13 Flower Development of Phalaenopsis Orchids Involves Functionally Divergent B-Class and E-Class MADS-Box Genes
- Chapter 14 The Function of C/D-Class MADS Box Genes in Orchid Gynostemium and Ovule Development
- Chapter 15 The Dual Function of Harpin in Disease Resistance and Growth in Phalaenopsis Orchids
- Chapter 16 Development of a High-Throughput Virus-Induced Gene-Silencing Vector in Phenotypic Screening of Orchids
- Chapter 17 Application of VIGS to Floral Gene Function Studies of the Orchid, a Nonmodel Plant
- Chapter 18 Functional Study of Phytoene Synthase by RNAi-Based Downregulation in the Oncidesa Orchid
- Chapter 19 Flower Color and Pigmentation Patterns in Phalaenopsis Orchids
- Chapter 20 Prolonged Exposure to Elevated Temperature Induces Floral Transition via Upregulation of Cytosolic Ascorbate Peroxidase 1 and Subsequent Reduction of the Ascorbate Redox Ratio in the Oncidium Hybrid Orchid 421 Dan-Chu Chin and Kai-Wun Yeh
- Chapter 21 Methylation Effect on Chalcone Synthase Gene Expression Determines Anthocyanin Pigmentation in Floral Tissues of Two Oncidium Orchid Cultivars
- Index