
Metalloprotein Active Site Assembly
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Metalloprotein Active Site Assembly
About this book
Summarizes the essential biosynthetic pathways for assembly of metal cofactor sites in functional metalloproteins
Metalloprotein Active Site Assembly focuses on the processes that have evolved to orchestrate the assembly of metal cofactor sites in functional metalloproteins. It goes beyond the simple incorporation of single metal ions in a protein framework, and includes metal cluster assembly, metal-cofactor biosynthesis and insertion, and metal-based post-translational modifications of the protein environments that are necessary for function. Several examples of each of these areas have now been identified and studied; the current volume provides the current state-of-the-art understanding of the processes involved.
An excellent companion to the earlier book in this series Metals in Cells—which discussed both the positive and negative effects of cellular interactions with metals—this comprehensive book provides a diverse sampling of what is known about metalloprotein active site assembly processes. It covers all major biological transition metal components (Mn, Fe, Co, Ni, Mo), as well as the other inorganic components, metal-binding organic cofactors (e.g., heme, siroheme, cobalamin, molybdopterin), and post-translationally modified metal binding sites that make up the patchwork of evolved biological catalytic sites. The book compares and contrasts the biosynthetic assembly of active sites involving all biological metals. This has never been done before since it is a relatively new, fast-developing area of research.
Metalloprotein Active Site Assembly is an ideal text for practitioners of inorganic biochemistry who are studying the biosynthetic pathways and gene clusters involved in active site assembly, and for inorganic chemists who want to apply the concepts learned to potential synthetic pathways to active site mimics.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
Part 1
Assembly and Trafficking of Simple Fe-S Clusters
Nif System for Simple [Fe–S] Cluster Assembly in Nitrogen-Fixing Bacteria
- 1 Introduction
- 2 The Nif System as a Model for Analysis of Simple [Fe–S] Cluster Assembly
- 3 Analysis of the Mo-Dependent Nitrogen-Fixing System in A. vinelandii
- 4 Genetic Phenotypes and Biochemical Features Indicated a Role for NifU and NifS in [Fe–S] Cluster Formation
- 5 NifS Cysteine Desulfurase
- 6 NifU Provides a Scaffold for [Fe–S] Cluster Assembly
- 7 NifU and NifS as the Minimum Set for the Assembly and Transfer of Fe–S Clusters
- 8 The NifS/NifU [Fe–S] Cluster Assembly Toolkit Provides a Paradigm for Simple [Fe–S] Cluster Assembly
- 9 Functional Cross Talk Between [Fe–S] Cluster Biosynthetic Systems
- 10 Concluding Remarks
- 11 Acknowledgments
- 12 Related Articles
- 13 Abbreviations and Acronyms
- 14 References
1 Introduction
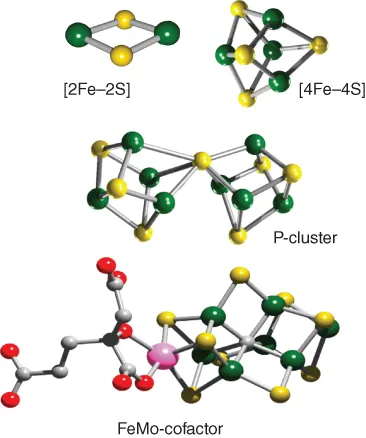
2 The Nif System as a Model for Analysis of Simple [Fe–S] Cluster Assembly
Table of contents
- Cover
- Title Page
- Copyright
- Table of Contents
- Encyclopedia of Inorganic and Bioinorganic Chemistry
- Contributors
- Series Preface
- Volume Preface
- Periodic Table of the Elements
- Part 1: Assembly and Trafficking of Simple Fe-S Clusters
- Part 2: Assembly of Complex and Heterometallic Fe-S Cluster Active Sites
- Part 3: Assembly of Homometallic and Heterometalic Cu Cluster Active Sites
- Part 4: Assembly of Homometallic and Heterometallic Mn Clusters
- Part 5: Assembly of Homometallic and Heterometallic Ni Clusters
- Part 6: Assembly of Cofactors for Binding Active-site Metal Centers
- Index
- Abbreviations and Acronyms used in this Volume
- End User License Agreement