
Evolutionary Computation in Gene Regulatory Network Research
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Evolutionary Computation in Gene Regulatory Network Research
About this book
Introducing a handbook for gene regulatory network research using evolutionary computation, with applications for computer scientists, computational and system biologists
This book is a step-by-step guideline for research in gene regulatory networks (GRN) using evolutionary computation (EC). The book is organized into four parts that deliver materials in a way equally attractive for a reader with training in computation or biology. Each of these sections, authored by well-known researchers and experienced practitioners, provides the relevant materials for the interested readers. The first part of this book contains an introductory background to the field. The second part presents the EC approaches for analysis and reconstruction of GRN from gene expression data. The third part of this book covers the contemporary advancements in the automatic construction of gene regulatory and reaction networks and gives direction and guidelines for future research. Finally, the last part of this book focuses on applications of GRNs with EC in other fields, such as design, engineering and robotics.
• Provides a reference for current and future research in gene regulatory networks (GRN) using evolutionary computation (EC)
• Covers sub-domains of GRN research using EC, such as expression profile analysis, reverse engineering, GRN evolution, applications
• Contains useful contents for courses in gene regulatory networks, systems biology, computational biology, and synthetic biology
• Delivers state-of-the-art research in genetic algorithms, genetic programming, and swarm intelligence
Evolutionary Computation in Gene Regulatory Network Research is a reference for researchers and professionals in computer science, systems biology, and bioinformatics, as well as upper undergraduate, graduate, and postgraduate students.
Hitoshi Iba is a Professor in the Department of Information and Communication Engineering, Graduate School of Information Science and Technology, at the University of Tokyo, Toyko, Japan. He is an Associate Editor of the IEEE Transactions on Evolutionary Computation and the journal of Genetic Programming and Evolvable Machines.
Nasimul Noman is a lecturer in the School of Electrical Engineering and Computer Science at the University of Newcastle, NSW, Australia. From 2002 to 2012 he was a faculty member at the University of Dhaka, Bangladesh. Noman is an Editor of the BioMed Research International journal. His research interests include computational biology, synthetic biology, and bioinformatics.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
II
EAs FOR GENE EXPRESSION DATA ANALYSIS AND GRN RECONSTRUCTION
CHAPTER 4
BICLUSTERING ANALYSIS OF GENE EXPRESSION DATA USING EVOLUTIONARY ALGORITHMS
Griffith University, Queensland, Australia
4.1 INTRODUCTION
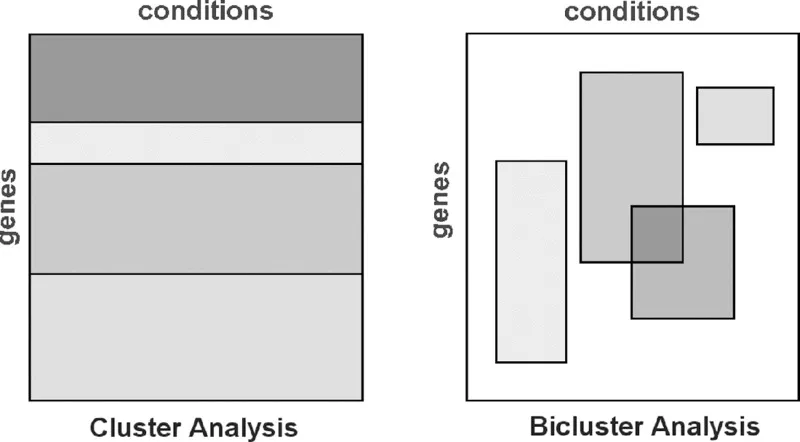
Table of contents
- Cover
- Series
- Title page
- Copyright
- PREFACE
- ACKNOWLEDGMENTS
- CONTRIBUTORS
- I PRELIMINARIES
- II EAs FOR GENE EXPRESSION DATA ANALYSIS AND GRN RECONSTRUCTION
- III EAs FOR EVOLVING GRNs AND REACTION NETWORKS
- IV APPLICATION OF GRN WITH EAs
- INDEX
- SERIES
- EULA
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app