Hydrogen Exchange Mass Spectrometry of Proteins
Fundamentals, Methods, and Applications
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Hydrogen Exchange Mass Spectrometry of Proteins
Fundamentals, Methods, and Applications
About this book
Hydrogen exchange mass spectrometry is widely recognized for its ability to probe the structure and dynamics of proteins. The application of this technique is becoming widespread due to its versatility for providing structural information about challenging biological macromolecules such as antibodies, flexible proteins and glycoproteins. Although the technique has been around for 25 years, this is the first definitive book devoted entirely to the topic.
Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods and Applications brings into one comprehensive volume the theory, instrumentation and applications of Hydrogen Exchange Mass Spectrometry (HX-MS) - a technique relevant to bioanalytical chemistry, protein science and pharmaceuticals. The book provides a solid foundation in the basics of the technique and data interpretation to inform readers of current research in the method, and provides illustrative examples of its use in bio- and pharmaceutical chemistry and biophysics
In-depth chapters on the fundamental theory of hydrogen exchange, and tutorial chapters on measurement and data analysis provide the essential background for those ready to adopt HX-MS. Expert users may advance their current understanding through chapters on methods including membrane protein analysis, alternative proteases, millisecond hydrogen exchange, top-down mass spectrometry, histidine exchange and method validation. All readers can explore the diversity of HX-MS applications in areas such as ligand binding, membrane proteins, drug discovery, therapeutic protein formulation, biocomparability, and intrinsically disordered proteins.
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
1
Hydrogen Exchange: A Sensitive Analytical Window into Protein Conformation and Dynamics
1.1 Isotopic Exchange and the Study of Protein Conformation and Dynamics
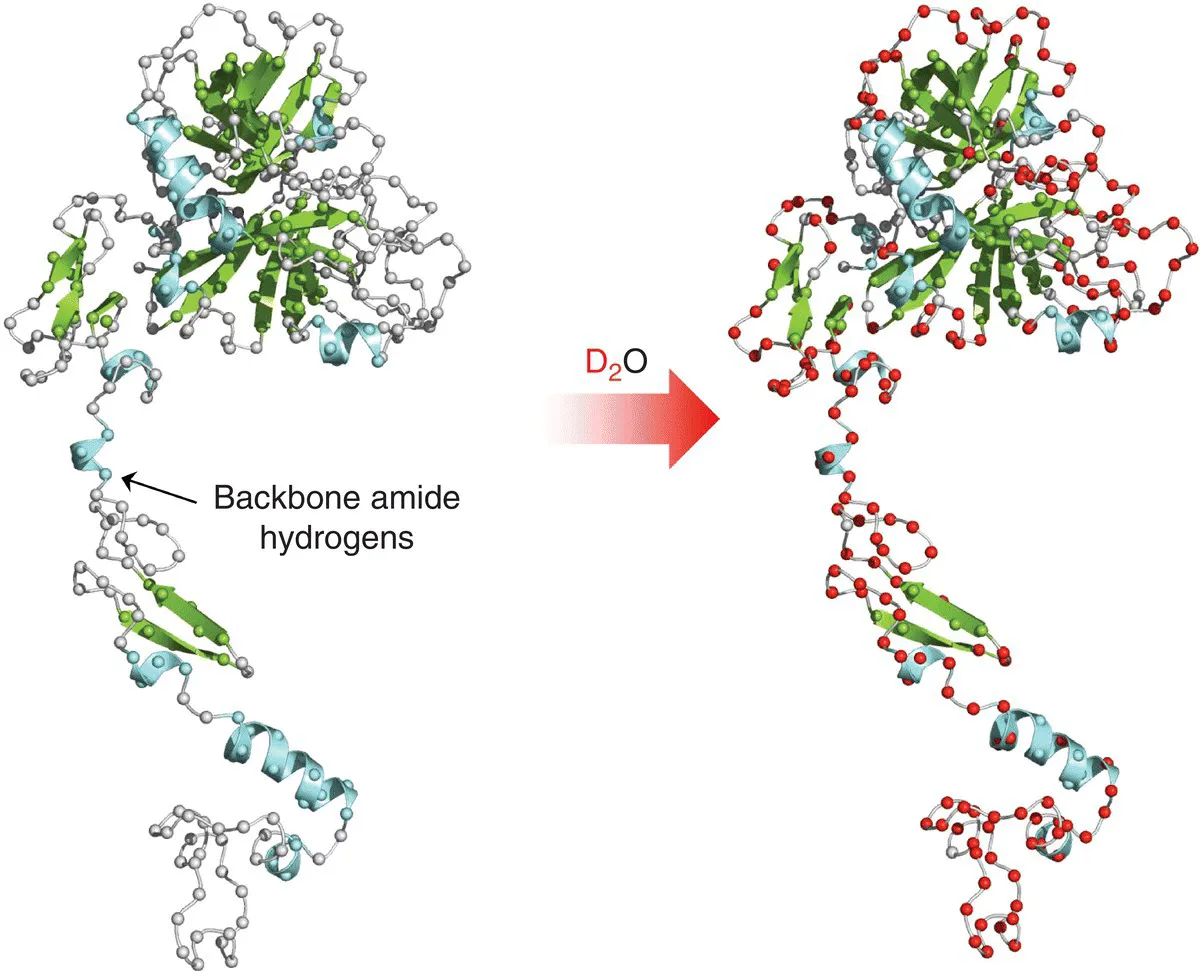
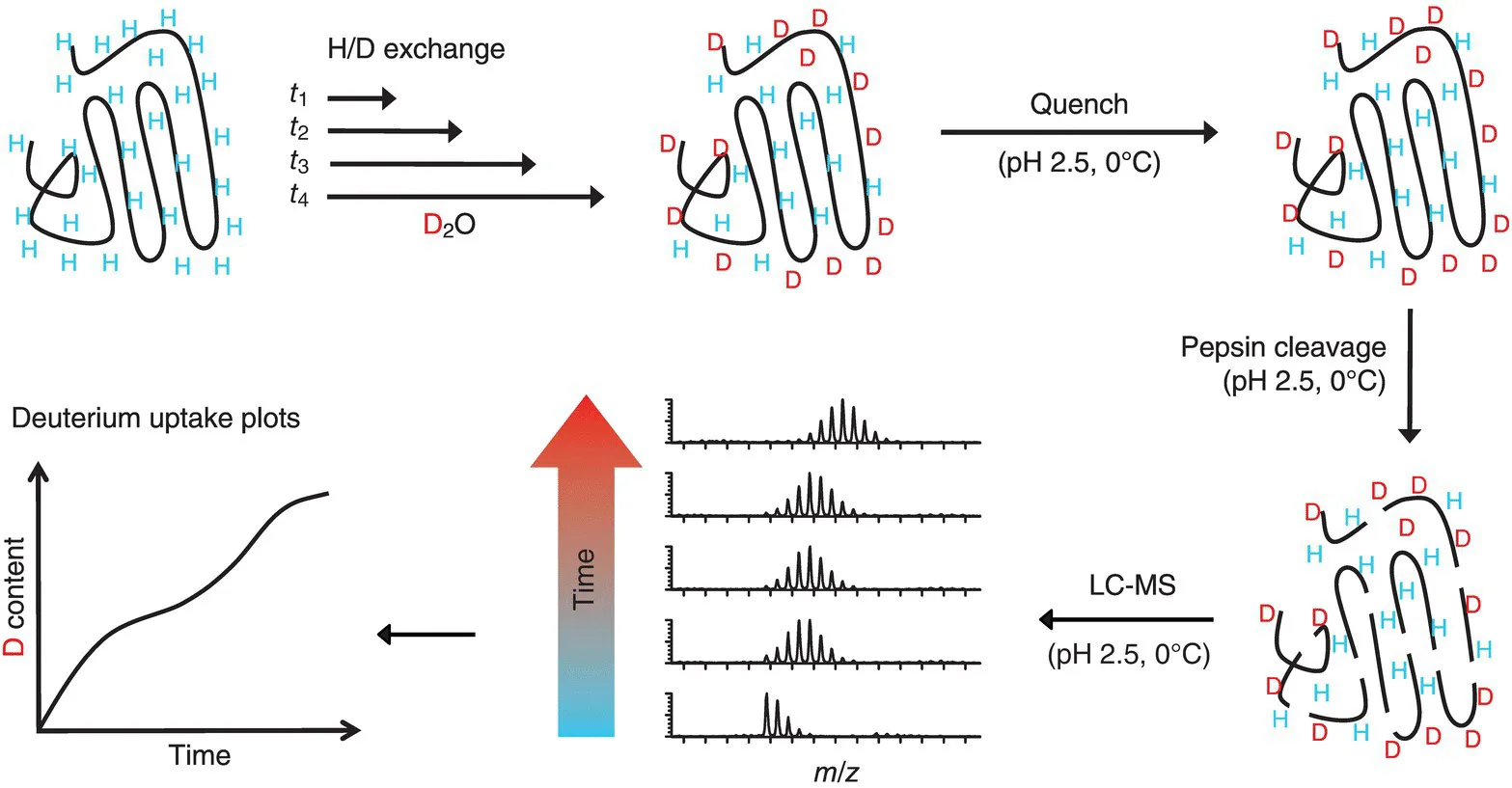
Table of contents
- Cover
- Title Page
- Table of Contents
- List of Contributors
- Foreword
- Preface
- A Note about Nomenclature
- 1 Hydrogen Exchange
- 2 Hydrogen Exchange Mass Spectrometry Experimental Design
- 3 Data Processing in Bottom-Up Hydrogen Exchange Mass Spectrometry
- 4 Method Validation and Standards in Hydrogen Exchange Mass Spectrometry
- 5 Millisecond Hydrogen Exchange
- 6 Proteases for Hydrogen Exchange Mass Spectrometry
- 7 Extracting Information from Hydrogen Exchange Mass Spectrometry Data
- 8 Gas-Phase Fragmentation of Peptides to Increase the Spatial Resolution of the Hydrogen Exchange Mass Spectrometry Experiment
- 9 Top-Down Hydrogen Exchange Mass Spectrometry
- 10 Histidine Hydrogen Exchange for Analysis of Protein Folding, Structure, and Function
- 11 Hydrogen Exchange Mass Spectrometry for the Analysis of Ligand Binding and Protein Aggregation
- 12 Application of Differential Hydrogen Exchange Mass Spectrometry in Small Molecule Drug Discovery
- 13 The Role of Hydrogen Exchange Mass Spectrometry in Assessing the Consistency and Comparability of the Higher-Order Structure of Protein Biopharmaceuticals
- 14 Utility of Hydrogen Exchange Mass Spectrometry in Epitope Mapping
- 15 Hydrogen Exchange Mass Spectrometry for Proteins Adsorbed to Solid Surfaces, in Frozen Solutions, and in Amorphous Solids
- 16 Hydrogen Exchange Mass Spectrometry of Membrane Proteins
- 17 Analysis of Disordered Proteins by Hydrogen Exchange Mass Spectrometry
- 18 Hydrogen Exchange Mass Spectrometry as an Emerging Analytical Tool for Stabilization and Formulation Development of Therapeutic Monoclonal Antibodies
- Index
- End User License Agreement