![]()
1
Applications of Nutrigenomics for Disease Prevention and Better Health
Adnan Ali and Yashwant V. Pathak
University of South Florida
Tampa, Florida
Ali M. Ardekani
Middle-Eastern Association for Cancer Research (MEACR)
Montreal, Québec, Canada
Contents
1.1 Nutrigenomics in the genomics and postgenomics age
1.2 Nutrigenomics: A brief introduction
1.3 A primer on genomics and beyond
1.3.1 Genome-wide association studies
1.3.2 Transcriptomics
1.3.3 Epigenomics
1.4 Molecular biology tools for gene editing
1.4.1 Obesity and the FTO locus
1.5 The microbiome
1.6 Conclusion
References
1.1 Nutrigenomics in the genomics and postgenomics age
The study of nutrition has long preceded the genomics age. Humans, since time immemorial, have been fascinated by how the nutrients we intake influence our health. In the late 1990s and early 2000s, a fundamental shift occurred in nutritional research. Many nutritional researchers, realizing that a fundamental understanding of how nutrition impacts health cannot be achieved without a thorough understanding of how nutritional pathways work on a molecular and genetic level, began to leverage new technologies and experimental data collected from the burgeoning age of sequencing (Müller and Kersten 2003). With the ever-decreasing cost of DNA sequencing and other molecular biology advances, scientists have now gained an unprecedented ability to not only investigate how genetic variation impacts nutrition and health at a population level but also to dissect the basic molecular mechanisms and systems that integrate to regulate human nutrition. To gain a coherent understanding of nutrigenomics, one must first grasp the basic molecular tools and techniques that are used in modern-day genomics. Here, we will conduct a brief survey of the techniques and theories that underlie modern nutrigenomics. We will then illustrate the powerful uses of these tools through several case studies.
1.2 Nutrigenomics: A brief introduction
Nutrigenomics differs from traditional nutritional studies not only by the genomics-based methods by which it answers questions but also by integrating data from various systems to gain an integrative understanding of the gene–nutrition–disease associations at a mechanistic level, with the overall goal of creating personalized or adaptive nutritional regimes to promote better health and prevent disease. Therefore, nutrigenomics is concerned with both how nutrients impact gene expression or structure and how genetic differences impact the way nutrient response happens. To understand how genetics and nutrition interact, we must first discuss the impacts of the environment and genetics on phenotype. To start, one can write the following simple equation for the phenotype:
p = g + e
var (p) = var (g) + var (e) + 2 cov (g, e)
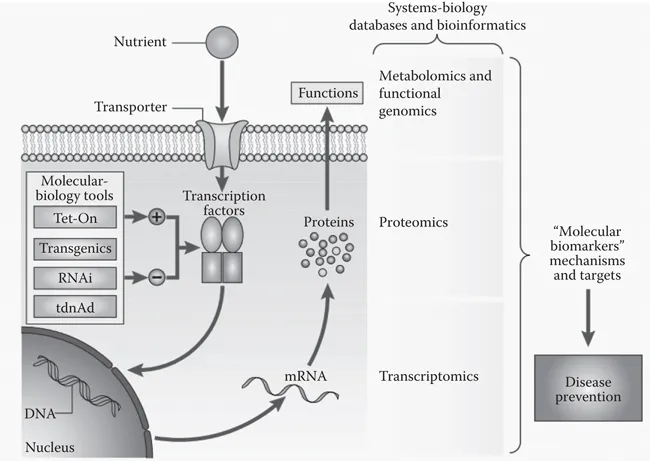
The first equation states that the phenotype (p) is the sum of genetic (g) and environmental (e) factors. If we want to learn about the phenotypic variability of an individual, we can partition the phenotypic variance into the sum of genetic variation, environmental variation, and covariation between genetics and the environment (Simopoulos, 2010). The notion that a certain proportion of the variance in phenotype can be determined by genetics is encapsulated in the concept of heritability, which we can calculate for any phenotype. Heritability can be measured in several different ways, including broad-sense heritability, which is (Hartl et al. 1997). A narrower definition of heritability, known as narrow-sense heritability, only takes into account additive genetic effects and excludes effects from dominance and allele-to-allele effects like epistasis. Narrow-sense heritability is denoted as Nutrition itself is strictly an environmental effect, and its effects are captured by the portion of the phenotypic variability that is not heritable, in other words, (Hartl et al. 1997). It is perhaps illustrative to think about human height, a phenotype that is determined by both genetics and environmental factors, such as nutritional intake. From a recent meta-analysis of many different height heritability studies, we currently estimate that the h2 of height is at 0.73 ± 0.03 (Polderman et al. 2015). This means that environmental factors and gene–environment interactions impact the remaining 0.27 proportion of phenotypic variance not explained by genetics, of which nutrition and the interaction between genetics and nutrition play a critical role. It is extremely difficult to partition the remaining 0.27 into the proportion that is assigned to environment alone or gene-to-environment interactions. Nutrigenomics itself is primarily concerned with the nutrition–gene interactions and their impact on various important phenotypes, such as disease phenotypes. Although many other systems, such as the microbiome and the epigenome, may change the model we have just postulated, it is important to keep in mind that, depending on the traits, the effects of nutrition on phenotype are primarily exerted at the environment and the gene–environmental interaction levels (Simopoulos, 2010). However, genes alone do not exert any effect. They must be transcribed to mRNA to form the transcriptome, and then they must be translated to proteins to form the proteome before exerting their effects on phenotype. As a result, nutrigenomics is also fundamentally interested in the effects of nutrition at the transcriptomic and proteomic levels as well. Indeed, nutrigenomics takes a systems-level approach to understanding the effects of nutrition on the human body (Astley 2007; Müller and Kersten 2003) (Figure 1.1).
Figure 1.1 The systems-level schema of nutrigenomics. Although a number of tools have changed since the time when this figure was created—such as CRISPR-based systems for gene editing—the overall schema of how nutrigenomics is integrated at the systems level has remained largely the same. (Reproduced from Müller, M. & Kersten, S., Nat. Rev. Genet., 4, 4, 315–322, 2003.)
By combining genomics, transcriptomics, proteomics, epigenomics, metabolomics, proteomics, and functional genomics with state-of-the-art bioinformatics tools and molecular-biology tools, modern nutrigenomics can not only identify the gene–nutritional interactions but also dissect the mechanistic foundations at a systems level.
1.3 A primer on genomics and beyond
The most basic level at which genetics can influence nutrition is the DNA level. Variation in our DNA—mostly in the form of single nucleotide mutations known as single nucleotide polymorphisms (SNPs)—plays a critical role in determining the phenotypic variability in the human population. With advances in sequencing technology, we are now able to interrogate the entire human genome in large cohorts to ascertain the extent of human genetic variation. Large consortium studies such as the HapMap Project and the 1,000 Genomes Project have yielded many insights into the overall diversity of mutations in the human population (Altshuler et al. 2010; Auton et al. 2015). A typical human differs from the reference human genome at about 4.1–5.0 million sites across the genome, affecting over 20 million base pairs, with 99.9% of these sites being SNPs. Only about 2,000 variants per genome are associated with any documented diseases, and only about 24–30 variants per genome are implicated in rare diseases (Auton et al. 2015). Variants themselves can be split between coding and noncoding variants, as coding variants lie directly in the coding regions of genes and may result in a conformational change of the protein, while noncoding variants may affect many other regulatory elements in the genome, including transcription factor binding sites or promoter regions. Assigning functional significance to coding variants is significantly easier than assigning functional impact to noncoding variants. Through resources made possible by the Exome Aggregation Consortium, we are now able to assign functional importance to nearly every coding variant observed in the human population (Lek et al. 2015). It is abundantly clear that many of these variants affect biological processes and pathways that are related to nutrition and especially gene–nutrition interactions. For example, rs671 is a classic missense SNP that produces a defective version of aldehyde dehydrogenase 2 (ALDH2), a key gene in the oxidative pathway of alcohol metabolism. Individuals with either one or two copies of the A-allele will suffer from facial flushing and severe hangovers (Takeuchi et al. 2011; Wang et al. 2013). Recent evidence has shown that rs671 affects more than just alcohol metabolism, as it has been significantly implicated (Sakiyama et al. 2016). The manner by which an individual can avoid the deleterious effects of this variant is simple: consume less alcohol. Indeed, it has been observed that individuals with this variant do in fact consume less alcohol, and when they do consume alcohol, the body is significantly less efficient at metabolizing alcohol (Takeuchi et al. 2011; Wang et al. 2013). This example involving ALDH2 is more of illustrative of nutrigenetics—the study of interactions between one gene and one nutrient; however, this term is now becoming increasingly merged together with nutrigenomics, as awareness grows that many nutritionally related genes are pleiotropic—one gene affecting multiple phenotypes—or that certain nutritionally related phenotypes are multigenic (Simopoulos 2010).
1.3.1 Genome-wide association studies
In order to ascerta...