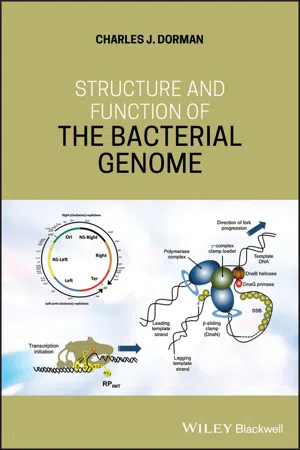
1.1 Genome Philosophy
The genome of a bacterium consists of its entire collective of genes, and these can be located in a chromosome (or chromosomes) and on extra‐chromosomal autonomous replicons such as plasmids. Chromosomes and plasmids replicate, copying the genes that they carry, with the replicon copies being segregated into the daughter cells at cell division. This process drives the vertical transmission of genetic information from one generation to the next and its fidelity determines the stability of the genetic information in the genome. If non‐lethal errors occur during the replication of the genome the resulting mutations will be transmitted to the next generation.
The vertical transmission of genetic copying errors is one of the driving forces of evolution in all types of organisms. Bacterial cells are also prone to the evolutionary influence that is horizontal gene transfer (HGT). Here, foreign genes are transmitted to the bacterium, adding to its genetic complement. Evolution through HGT is much less subtle than evolution through the vertical transmission of copying errors which often involves the gradual accumulation of single nucleotide changes to the genome over many generations. Other types of mutation that are transmitted vertically are inversions, insertions, and deletions of genomic DNA. In HGT a bacterium can acquire entire clusters of genes en bloc, resulting in the acquisition of novel capabilities in a single generation. Examples include the arrival of genes that make the bacterium resistant to an antibiotic or to a heavy metal that previously could kill it, or genes that allow the organism to colonise a niche in the environment from which it had previously been excluded.
HGT played a key role in the early research work that led to the mapping of the bacterial genome and to our understanding of the locations of its genes. Among the autonomously replicating plasmids found in bacteria are elements that can promote their own transfer from cell to cell. The fertility, or ‘F’, factor of Escherichia coli was among the first to be studied. F encodes proteins that can build a connecting bridge between the F+ (or male) cell and one that is F− (female). F transfers one of its DNA strands to the F− cell in a process called conjugation. This is the bacterial equivalent of sex. It resembles sex in higher organisms in that the participants are male (F+) and female (F−) but it differs from conventional sex because the process converts the female into an F+ male. The F plasmid has segments of DNA that are identical in sequence, or almost identical, to DNA segments found in the chromosome. Usually these are mobile genetic elements called insertion sequences (IS). The homologous recombination machinery of the cell can recombine the F‐associated region of DNA sequence identity with a chromosomal counterpart, causing F to become fused with that part of the chromosome (Hadley and Deonier 1980). Where this happens is determined by the location of the IS element, and these mobile elements can be found at sites distributed around the chromosome.
Once F has become one with the chromosome, it is replicated as a part of that molecule. It can still engage in conjugation, however. When this happens, the DNA that is transferred to the F− female bacterium consists of chromosomal DNA with F DNA in the vanguard. Strains that can act as DNA donors in these matings are called ‘Hfr’ (high frequency of recombination) (Reeves 1960) and homologous recombination between the incoming, horizontally transferred DNA and the resident chromosome allows the order of the genes on the chromosome to be determined. Experiments, in which chromosomal gene transfer mediated by the F plasmid was monitored as a function of time, allowed a rudimentary genetic map of the E. coli chromosome to be assembled (Bachmann 1983; Brooks Low 1991). Because the mating experiments were allowed to proceed for fixed periods of time before the deliberate breakage of the conjugation bridges by mechanical shearing, these early genetic maps were calibrated in ‘minutes’. It was discovered that it took 100 minutes to transfer the entire E. coli chromosome from one cell to another by conjugation (Bachmann 1983; Brooks Low 1991). Similar experiments were performed for other bacterial species, including the pathogen Salmonella, giving rough approximations of the physical scale of bacterial genomes (Sanderson and Roth 1988). Hfr strains could also mediate gene transfer between E. coli and Salmonella (Schneider et al. 1961). When the F plasmid is excised from the chromosome, genes that had been adjacent to the plasmid can be removed too, becoming part of the autonomously replicating episome. The plasmids are called F‐prime (F′) and have proved to be very useful in genetic analysis. The chromosomal gene ‘cargo’ can be transferred to F‐minus strains by conjugation and this phenomenon can be exploited in genetic complementation experiments. Work of this type provided useful information about gene order and the position and nature of genetic mutations. F‐primes have been used to investigate plasmid stability, incompatibility, and DNA replication: for example, the F′‐lac episome was used extensively to study plasmid replication in E. coli (Davis and Helmstetter 1973; Dubnau and Maas 1968). Experiments with E. coli mutants deficient in Hfr recombination led to the discovery of important genes involved in homologous recombination: for example, recA (Clark and Margulies 1965), recB, and recC (Barbour and Clark 1970; Willetts et al. 1969; Youngs and Bernstein 1973).
HGT also provided a means for more refined mapping of genomes. Bacteriophages (often abbreviated to ‘phages’) are viruses that replicate in bacterial cells. Some phages package bacterial DNA in their viral heads as they exit the bacterial host and this DNA is transferred to the next bacterium that they manage to infect in a process known as transduction. The length of the DNA segment that a phage head can accommodate is finite and known in the cases of the viruses most commonly used for generalised transduction in E. coli (P1, 100–115 kb) and Salmonella (P22, 42 kb) (Sternberg and Maurer 1991). Therefore, genes that are co‐transduced must be within a distance of one anothe...