
- 600 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Molecular Biology of B Cells
About this book
Molecular Biology of B Cells, Second Edition is a comprehensive reference to how B cells are generated, selected, activated and engaged in antibody production.All of these developmental and stimulatory processes are described in molecular, immunological, and genetic terms to give a clear understanding of complex phenotypes.Molecular Biology of B Cells, Second Edition offers an integrated view of all aspects of B cells to produce a normal immune response as a constant, and the molecular basis of numerous diseases due to B cell abnormality. The new edition continues its success with updated research on microRNAs in B cell development and immunity, new developments in understanding lymphoma biology, and therapeutic targeting of B cells for clinical application. With updated research and continued comprehensive coverage of all aspects of B cell biology, Molecular Biology of B Cells, Second Edition is the definitive resource, vital for researchers across molecular biology, immunology and genetics.- Covers signaling mechanisms regulating B cell differentiation- Provides information on the development of therapeutics using monoclonal antibodies and clinical application of Ab- Contains studies on B cell tumors from various stages of B lymphocytes- Offers an integrated view of all aspects of B cells to produce a normal immune response
Frequently asked questions
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app.
Information
The Structure and Regulation of the Immunoglobulin Loci
Abstract
Keywords
3D-structure; Antibody repertoire; B cell development; Combinatorial diversity; Epigenetic marking; Germline transcription; Immunoglobulin; Junctional diversity; Transcription factor; V(D)J recombination1. Introduction
2. Genomic Organization of the Mouse Immunoglobulin Heavy Chain Locus
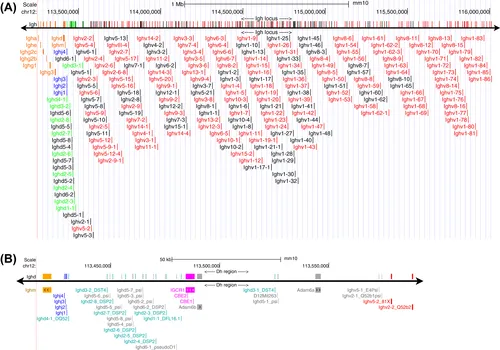
3. Genomic Organization of the Mouse Immunoglobulin Kappa Light Chain Locus
Table of contents
- Cover image
- Title page
- Table of Contents
- Copyright
- Dedication
- Preface
- Contributors
- Chapter 1. The Structure and Regulation of the Immunoglobulin Loci
- Chapter 2. The Mechanism of V(D)J Recombination
- Chapter 3. Transcriptional Regulation of Early B Cell Development
- Chapter 4. Relationships among B Cell Populations Revealed by Global Gene Analysis
- Chapter 5. Roles of MicroRNAs in B Lymphocyte Physiology and Oncogenesis
- Chapter 6. Proliferation and Differentiation Programs of Developing B Cells
- Chapter 7. Development and Function of B Cell Subsets
- Chapter 8. B Cells and Antibodies in Jawless Vertebrates
- Chapter 9. The Origin of V(D)J Diversification
- Chapter 10. Structure and Signaling Function of the B-Cell Antigen Receptor and Its Coreceptors
- Chapter 11. Fc and Complement Receptors
- Chapter 12. B Cell Localization and Migration in Health and Disease
- Chapter 13. B Cells as Regulators
- Chapter 14. B Cell Memory and Plasma Cell Development
- Chapter 15. The Role of the BAFF and Lymphotoxin Pathways in B Cell Biology
- Chapter 16. The Mucosal Immune System: Host–Bacteria Interaction and Regulation of Immunoglobulin A Synthesis
- Chapter 17. Gut Microbiota and Their Regulation
- Chapter 18. Molecular Mechanisms of AID Function
- Chapter 19. The Mechanism of IgH Class Switch Recombination
- Chapter 20. Somatic Hypermutation: The Molecular Mechanisms Underlying the Production of Effective High-Affinity Antibodies
- Chapter 21. Aberrant AID Expression by Pathogen Infection
- Chapter 22. Molecular Pathogenesis of B Cell Lymphomas
- Chapter 23. B Cells Producing Pathogenic Autoantibodies
- Chapter 24. The Cellular and Molecular Biology of HIV-1 Broadly Neutralizing Antibodies
- Chapter 25. Immune Deficiencies Caused by B Cell Defects
- Chapter 26. IMGT® Immunoglobulin Repertoire Analysis and Antibody Humanization
- Chapter 27. Anti-Interleukin-6 Receptor Antibody Therapy Against Autoimmune Inflammatory Diseases
- Chapter 28. Targeting the IL-17/IL-23 Axis in Chronic Inflammatory Immune-Mediated Diseases
- Chapter 29. Discovery and Development of Anti-TNF Therapy: Pillar of a Therapeutic Revolution
- Index