![]()
PART I
GENERAL ISSUES
![]()
1 FUNGI AND THEIR ALLIES
MEREDITH. BLACKWELL, JOSEPH W. SPATAFORA
A BROAD VIEW OF EUKARYOTES
KINGDOM FUNGI
PHYLUM CHYTRIDIOMYCOTA (ZOOSPORIC FUNGI)
PHYLUM ZYGOMYCOTA
CLADE GLOMALES
PHYLUM ASCOMYCOTA
Class Archiascomycetes
Class Saccharomycetes
Class Euascomycetes
PHYLUM BASIDIOMYCOTA
Class Ustilaginiomycetes
Class Urediniomycetes
Class Hymenomycetes
KINGDOM STRAMINIPILA (HETEROKONT ZOOSPORIC ORGANISMS)
Oomycota
Hyphochytriomycetes
Labyrinthulales and Thraustochytriales (and Aplanochytrium)
SLIME MOLDS
Plasmodiophorales (Parasitic Slime Molds)
Myxomycetes, Protostelids, and Dictyostelids (Plasmodial and Cellular Slime Molds)
Acrasid Slime Molds
CONCLUSIONS
Fungi are heterotrophic organisms that permeate our environment. With few exceptions fungi have filamentous bodies enclosed by cell walls, are nonmotile, and reproduce both sexually and asexually by spores. During the last decade, mycologists have made unprecedented progress toward producing a phylogenetic classification of fungi; a skeleton phylogeny based on analyses of DNA characters was developed relatively early on (Bruns et al. 1991; Alexopoulos et al. 1996; McLaughlin et al. 2001). Such a phylogeny serves as a basis on which hypotheses of fungal evolution can be developed. One important finding has been that “fungi” are polyphyletic (Table 1.1); their morphologies are convergent, having been derived independently from among several independent eukaryotic lineages (Fig. 1.1). A monophyletic group, exclusive of slime molds and oomycetes, is well defined and supported as “true fungi,” a kingdom-level taxon (Barr 1992; Bruns et al. 1993; Baldauf et al. 2000; Keeling et al. 2000).
TABLE 1.1 Fungi and Fungus-like Organisms Previously Classified as Fungi*
| Fungi |
| Chytridiomycota |
| Zygomycota |
| Ascomycota |
| Basidiomycota |
| Straminipila |
| Oomycota |
| Hyphochytriomycota |
| Labyrinthulales |
| Thraustochytriales |
| Slime molds |
| Plasmodiophorales |
| Myxomycota |
| Dictyosteliomycota |
| Acrasiomycota |
Historically fungi have been compared with plants and included in the study of botany. Contemporary studies, however, indicate that members of Kingdom Fungi are most closely related to animals, not plants, possibly through a choanoflagellate-like ancestor (Barr 1992; Baldauf and Palmer 1993; Wainright et al. 1993; Ragan et al. 1996; Baldauf et al. 2000; Cavalier-Smith 2001). The phylogenetic positions of other groups once considered as fungi are not always well defined among eukaryotes (see “Kingdom Straminipila” and “Slime Molds,” later in this chapter).
Thus far, fewer than 800 fungi or fungus-like taxa have been included together in any single phylogenetic reconstruction (Tehler et al. 2000), a number representing less than 1% of the 80,000 currently listed species (see Hawksworth et al. 1995). Estimates of up to 1.5 million species of fungi (six times more than the number of recognized species of land plants) are daunting; the missing fungi must be discovered, identified, and incorporated into the taxonomic framework. In addition to the relatively low numbers of taxa included in the trees, taxonomic coverage across higher taxa is limited. Until now only molecular characters from ribosomal RNA genes (rDNA) have been used widely in phylogenetic studies of fungi. The recent increase in use of other DNA regions and incorporation of phenotypic characters to resolve conflicting trees based on single genes and analytical approaches is encouraging.
Fungal classification, as opposed to phylogeny, is in a state of flux. Ideally, classifications should reflect phylogenetic relationships, but the relationships of all the groups covered in this volume have not been resolved nor have all of the groups been represented in analyses. Thus, the formal and complex classification systems that have been devised (e.g., Cavalier-Smith 2001) may vary in their inclusiveness. Systematists who study many types of organisms face this situation. Nevertheless, one reactionary suggestion called for a moratorium on publication of higher-level taxonomic systems until 2050 (Karatygin 1999). Hibbett and Donoghue (1998) suggested a more practical solution—the use of clade names rather than formal taxon designations. We use this approach, especially for higher taxa, in the following discussion.
Nowhere are examples of convergent evolution more evident than among organisms occupying the same habitat, which are grouped together for treatment in this volume. In this chapter we review the current state of knowledge of the phylogeny of true fungi (Kingdom Fungi) and fungus-like organisms placed in several additional distinct clades (Table 1.1). The two-volume work edited by McLaughlin et al. (2001) provides detailed information on fungi and fungus-like organisms; only a few of the chapters are cited here.
A BROAD VIEW OF EUKARYOTES
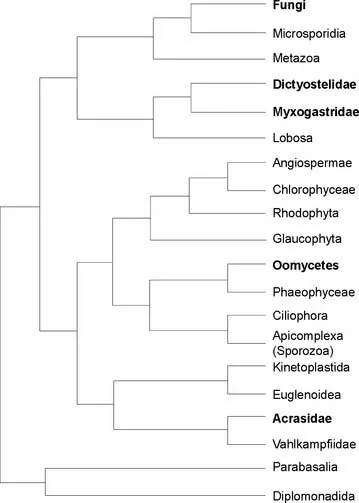
Previous studies based on small subunit rRNA genes (SSU rDNA) suggest that it may not be possible to reconstruct patterns of an ancient rapid radiation of the so-called “crown eukaryotes” (see “Kingdom Fungi,” later in this chapter), especially when based solely on characters from rDNA. Baldauf and colleagues (Baldauf et al. 2000), in contrast, used multiple genes and partitioning of data to develop a hypothesis of the relationships of the large diverse group of eukaryotes, which includes fungi and fungus-like organisms (Fig. 1.1). Their study differed from others in its approach because Baldauf and colleagues (2000) analyzed deduced amino acid sequences from four protein-coding regions (α-tubulin, β-tubulin, actin, and elongation factor 1-alpha [EF-1α]) in addition to rDNA. Consequently, they obtained better resolution of the deep branches in the tree (that is, those that represent the earliest divergences over a great period).
Many of the relationships proposed in Baldauf and colleagues’ (2000) study were not entirely new; however, the large data set they used supported several controversial or conflicting tree branches, depending on their use of either rDNA or protein-coding genes. Relationships identified or strengthened in their phylogenetic analyses included the Fungi-Microsporidia link, the grouping of dictyostelid and plasmodial slime molds, and the remote basal position of the acrasid slime molds. In some instances, correspondence of non-molecular traits (e.g., flagellum type) and arrangement of mitochondrial cristae provided additional support. Taking a broad view of fungi and fungus-like organisms and using a wide variety of characters, Baldauf and colleagues (2000) derived the following relationships (Fig. 1.1):
• Kingdom Fungi, (traditionally the phyla Ascomycota, Basidiomycota, Zygomycota, and Chytridiomycota; see “Kingdom Fungi,” later in this chapter) comprises a monophyletic group of “true” Fungi.
• Microsporidia are a sister group to Fungi, and these two taxa form a monophyletic grouping with the Metazoa (animals) linked by a choanoflagellate-like common ancestor.
• Although still scantily sampled, dictyostelid...