Protein Bioinformatics
From Sequence to Function
M. Michael Gromiha
- 339 pagine
- English
- ePUB (disponibile sull'app)
- Disponibile su iOS e Android
Protein Bioinformatics
From Sequence to Function
M. Michael Gromiha
Informazioni sul libro
One of the most pressing tasks in biotechnology today is to unlock the function of each of the thousands of new genes identified every day. Scientists do this by analyzing and interpreting proteins, which are considered the task force of a gene. This single source reference covers all aspects of proteins, explaining fundamentals, synthesizing the latest literature, and demonstrating the most important bioinformatics tools available today for protein analysis, interpretation and prediction. Students and researchers of biotechnology, bioinformatics, proteomics, protein engineering, biophysics, computational biology, molecular modeling, and drug design will find this a ready reference for staying current and productive in this fast evolving interdisciplinary field.
- Explains all aspects of proteins including sequence and structure analysis, prediction of protein structures, protein folding, protein stability, and protein interactions
- Presents a cohesive and accessible overview of the field, using illustrations to explain key concepts and detailed exercises for students.
Domande frequenti
Informazioni
Proteins
Publisher Summary
1.1 Building blocks
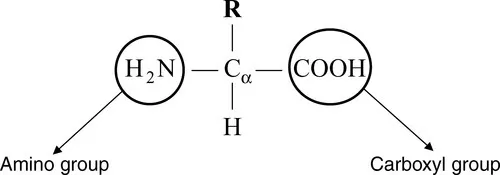
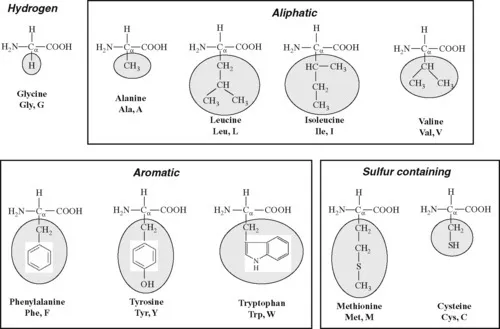
1.1.1 Amino acids
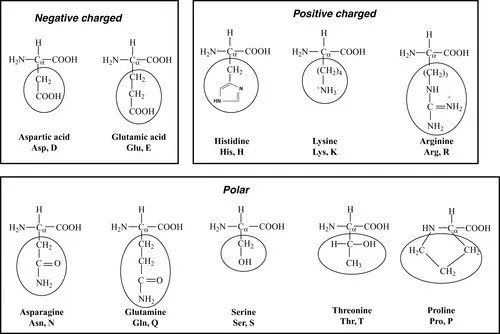
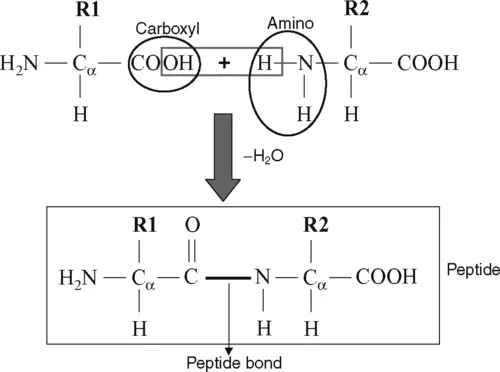
1.1.2 Formation of peptide bonds