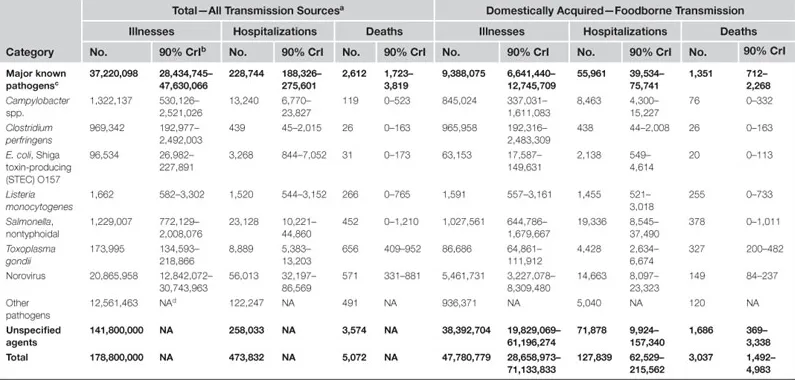
Data from surveillance, surveys, and other sources were used to estimate the number of domestically acquired foodborne illnesses, hospitalizations, and deaths caused by 31 major known pathogens, including 21 bacterial, 5 viral, and 5 parasitic pathogens (see Table 1.1) [6]. These known pathogens were estimated to cause 9.4 million (90% credible interval [CrI]: 6.6–12.7 million) domestically acquired foodborne illnesses, 55,961 hospitalizations (90% CrI: 39,534–75,741), and 1351 deaths (90% CrI: 712–2268) each year. Norovirus was estimated to cause the most foodborne illness (58%), while nontyphoidal Salmonella spp. was the leading cause of hospitalization (35%) and death (28%). Seven pathogens—Campylobacter spp., Clostridium perfringens, E. coli O157, Listeria monocytogenes, nontyphoidal Salmonella spp., norovirus, and Toxoplasma gondii—were estimated to cause 90% of domestically acquired foodborne illnesses, hospitalizations, and deaths due to the major known pathogens.
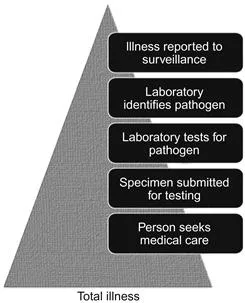
Estimating illness using the “burden-of-illness pyramid”
Most known pathogens had laboratory-based surveillance data available; therefore, the total number of illnesses was estimated using the “burden-of-illness pyramid” approach (Figure 1.1). Several steps are necessary for an illness to be included in laboratory-based surveillance: the ill person must seek medical care, a specimen must be submitted for laboratory testing, the laboratory must test for and identify the causative agent, and the illness must be reported to the public health authorities. Estimating the frequency of cases of foodborne disease that are not reported to public health from laboratories provides insight into under-reporting in the surveillance system. Similarly, assessing differences in medical care–seeking behavior, specimen submission, laboratory testing, or laboratory test sensitivity characterizes under-diagnosis at each step of the surveillance system. Accounting for the proportion of cases missed in traditional surveillance due to under-diagnosis and under-reporting builds the “burden-of-illness pyramid”. This allows for an extrapolation from laboratory-confirmed illnesses (at the top of the “burden-of-illness pyramid”) to estimate the overall number of illnesses in the community (at the bottom of the “burden-of-illness pyramid”). To extrapolate, a multiplier, the inverse of a proportion, is calculated for each surveillance step. For example, if the laboratory test sensitivity of a particular pathogen was estimated to be 80%, the multiplier for this surveillance step would be 1.25 (i.e., for every case of infection diagnosed an estimated 1.25 cases would have been tested for that pathogen).
Figure 1.1 Surveillance steps that must occur for laboratory-confirmed cases to be reported to surveillance.
In the United States, data on laboratory-confirmed illnesses caused by 25 of the 31 known pathogens were available from one or more of five surveillance systems: the foodborne diseases active surveillance network (FoodNet), the national notifiable disease surveillance system (NNDSS), the cholera and other vibrio illness surveillance (COVIS) system, the national tuberculosis surveillance system (NTSS), and the foodborne disease outbreak surveillance system (FDOSS). Similar to other countries, laboratory-based surveillance systems in the United States rely largely upon passive reports of diseases from clinical laboratories to state and local health departments, which are, in turn sent to the CDC. To assure that all laboratory-confirmed cases occurring within the FoodNet surveillance area are reported, personnel actively contact all laboratories in the catchment area. Therefore, when data were available in more than one surveillance system, active surveillance data from FoodNet were used, except for Vibrio spp., for which COVIS was used because of geographical clustering of Vibrio infections outside the FoodNet sites. Data on outbreak-associated illnesses from FDOSS were used only for pathogens with no data available from the other systems due to not being specifically reported or only manifesting as outbreaks.
Because FoodNet conducts active surveillance, the pathogens under FoodNet surveillance were assumed to have no under-reporting. Because COVIS and NNDSS are passive surveillance systems, an under-reporting multiplier (1.1 for bacterial and 1.3 for parasitic pathogens), derived by comparing the incidence of all nationally notifiable illnesses ascertained through FoodNet with that reported to NNDSS, was applied to those pathogen counts. For the five bacterial pathogens for which only outbreak data were available, an outbreak under-reporting multiplier was created by determining the proportion of illnesses in FoodNet caused by Campylobacter, Cryptosporidium, Cyclospora, Listeria, Salmonella, Shigella, Shiga toxin-producing E. coli (STEC), Vibrio, and Yersinia that were also reported as outbreaks associated to FDOSS. It was assumed that all Mycobacterium bovis illnesses were reported to NTSS.
To adjust for medical care seeking and specimen submission, the proportion of persons reporting an acute diarrheal illness (defined as ≥3 loose stools in a 24-hour period and lasting longer than one day or resulting in restricted daily activities) in the past month who sought medical care and submitted a stool sample for that illness were estimated using data from FoodNet surveys of the general population (FoodNet Population Surveys). Because persons with more severe illness are more likely to seek care [7], the rate of medical care seeking and stool sample submission was estimated separately for persons with bloody and non-bloody diarrhea; these proportions were used as surrogates for severe and mild presentations of most illnesses. These multipliers were derived by examining data on the proportion of patients with diarrhea seeking care and submitting specimens with different symptom profiles from population-based surveys. Multipliers for medical care seeking and stool sample submission (for those with mild and severe illness) were then appli...