Biological Sciences
Protein Structure
Protein structure refers to the specific three-dimensional arrangement of atoms within a protein molecule. This structure is crucial for the protein's function and can be described at different levels, including primary, secondary, tertiary, and quaternary structures. The unique folding and interactions between amino acids determine the protein's shape and ultimately its biological activity.
Written by Perlego with AI-assistance
Related key terms
1 of 5
11 Key excerpts on "Protein Structure"
- eBook - PDF
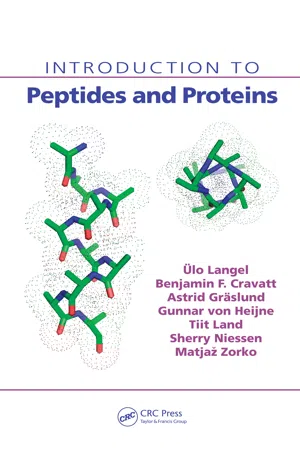
- Ulo Langel, Benjamin F. Cravatt, Astrid Graslund, N.G.H. von Heijne, Matjaz Zorko, Tiit Land, Sherry Niessen(Authors)
- 2009(Publication Date)
- CRC Press(Publisher)
The primary structure: the linear amino acid sequence of the polypeptide chain, including posttranslational modifications and disulfide bonds. 2. The secondary structure: the local structure of the linear segments of the polypeptide backbone atoms without regard to the conformation of the side chains. This level includes two sublevels: the super-secondary structure and the domains. The super-secondary structure means the association of the local secondary structural elements through side-chain interactions. The elements of the super-secondary structure are also called the motifs. A domain is usually a larger part of the protein sequence that can function and exist independently of the rest of the protein chain. 3. The tertiary structure: the three-dimensional arrangement of all the atoms in a polypeptide chain. In the single-chain proteins, this structure is functional. 4. The quaternary structure: the arrangement of the separate polypeptide chains (subunits) into the multi-subunit functional protein. 3.1 THE PRIMARY STRUCTURE The primary structure of a protein is the exact sequence of the amino acids joined by the peptide bond in a given polypeptide. In multichain proteins, each chain is referred to separately. By convention the primary structure is reported using three-letter or one-letter amino-acid coding (see Chapter 1), starting from the aminoter-minal (N) end and finishing at the carboxyl-terminal (C) end. The primary structure also requires specifying the crosslinking cysteines involved in the protein’s disul-fide bonds as well as all the posttranslational modifications in the polypeptide chain (see Chapter 5). Frequently, the modifications include cleavage of the polypeptide chain; therefore, the sequence of the polypeptide does not always correspond to the sequence of its mature mRNA. The linear polypeptide chain folds in a particular arrangement, giving a defined three-dimensional structure. - eBook - PDF
- Bhupendra Pushkar(Author)
- 2019(Publication Date)
- Delve Publishing(Publisher)
Analysis of Protein Structure 3 CONTENTS 3.1. Introduction ...................................................................................... 38 3.2. Crystallography ................................................................................. 44 3.3. Modeling .......................................................................................... 51 Protein Bioinformatics 38 3.1. INTRODUCTION To comprehend the fundamental standards of protein three-dimensional structure and the capability of their utilization in different regions of research, scholastic, or modern like pharmaceutical or biotech businesses. We first need to take a gander at the four degrees of Protein Structure. The distinctive secondary levels rely upon one another, together making an amazingly unpredictable system of communications somewhere in the range of thousands of iotas. The primary level is the amino acid sequence alignment – there are 20 diverse amino acids most usually found in proteins. The succession of these amino acids in a polypeptide chain basically decides the kinds of secondary structure components present, which is the second degree of association and the route by which they are masterminded in space, making basic themes and rows, which is the third degree of association. A free collapsing unit of the three-dimensional Protein Structure is known as a space. It is autonomous in light of the fact that spaces may frequently be cloned, communicated and refined freely of the remainder of the protein, and they may even show action, if there is any realized movement related with them. A few proteins contain one single space while others may contain a few areas. A protein space is appointed a particular kind of raw. Areas with a similar overlay could possibly be identified with one another practically. This is essentially on the grounds that Nature has re-utilized a similar overlap commonly in various settings. - eBook - PDF
- BIOTOL, B C Currell, R C E Dam-Mieras(Authors)
- 2013(Publication Date)
- Butterworth-Heinemann(Publisher)
This overall conformation is the consequence of the presence of any secondary structures and how they and the remainder of the chain are positioned. Each protein adopts a unique tertiary structure. This conformation will be the result of the various weak bonds which can arise. It also depends on interaction with the surrounding solvent (usually water). Thus the unique shape of a protein is that of lowest free energy, which is achieved by minimising unfavourable interactions (eg hydrophobic groups in contact with water, where they disrupt hydrogen bonding between water molecules) whilst maximising favourable interactions. Myoglobin was the first protein whose structure was determined by X-ray diffraction analysis. Myoglobin is the oxygen-storing protein of muscle and is relatively small (mol weight= 17500 daltons: 153 amino acids), yet it represented a breakthrough in biochemistry. Subsequently, larger proteins and those with subunits (eg haemoglobin) were described. The complete structure of an enzyme (lysozyme) enabled a detailed description of the catalytic mechanism by which an enzyme accelerates reaction rates to be convincingly proposed. Our detailed understanding of the structure and action of proteins relies on application of X-ray diffraction. So what did the X-ray diffraction analysis of myoglobin reveal? Myoglobin was shown (Figure 3.16) to be very compact, with water almost completely excluded from the interior of the protein. It is made up of 8 sections of a-helix (three-quarters of all residues are present in -helices), which fold to make up a box-like molecule; proline forces a bend at the end of some helices. Strikingly, the interior of the molecule contains almost exclusively hydrophobic, non-polar residues, tightly packed together. Almost all the polar residues are on the surface of the molecule, where the sidechains interact favourably with water (via hydrogen bonding). - eBook - PDF
- Bhupendra Pushkar(Author)
- 2020(Publication Date)
- Delve Publishing(Publisher)
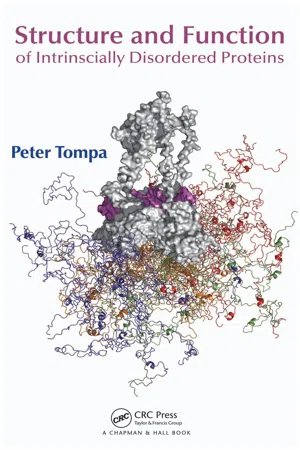
In fact, in cellular DNA, each gene tends to contain the code for any particular Protein Structure. These proteins are assembled with different amino acid sequences. Along with this, they also are combined together by many different bonds and folded into various three-dimensional structures. The folded shape or conformation relies on the linear amino acid sequence of the protein directly. The shape of a protein is critical to its function. The reason behind this is that it determines whether the proteins can interact with some other molecules. The Protein Structures are considered to be very complex and therefore, the researchers have gained the ability to easily as well as quickly determine the structure of the complete proteins down to the atomic level. There are techniques that are used in the period of the 1950s but now in today’s era, they were very slow as well as laborious to be used. And this is why the complete Protein Structures were very slow to be solved. Conceptually, the early structural biochemists divided the Protein Structures into four levels to make it easier to get a handle on the complexity of the entire structures. It is important to understand these four levels of Protein Structure named as primary, secondary, tertiary and quaternary in order to determine the way in which the protein gets its final shape or conformation. The primary structure of a protein is the unique sequence of the amino acids in every polypeptide chain which makes up the protein. This is just several amino acids that appear in the order in a polypeptide chain and it is not really a structure. This is referred to as the primary structure of the polypeptide chain because of the fact that the final Protein Structure ultimately depends on this sequence. An example can be the pancreatic hormone insulin which has two polypeptide chains namely A and B. - Peter Tompa, Alan Fersht(Authors)
- 2009(Publication Date)
- Chapman and Hall/CRC(Publisher)
1 1 Principles of Protein Structure and Function The principles of Protein Structure surveyed in this chapter have been established mostly by studying globular proteins. The structure of a globular protein can be described by the coordinates of all its atoms, but this information is often too complex to interpret in terms of function. Thus, scientists have devised a hierarchical vocabulary that can describe different levels of structure from the sequence of amino acids to the spatial arrangement of subunits. Further levels of complexity, such as post-translational modifications, the process of acquiring the 3-D structure (folding), and the description of the unfolded state resembling intrinsically disordered proteins (IDPs), also pertain to the comprehensive structural description of proteins. It has long been thought that the description of (ordered) proteins by these concepts provides a universal key to understanding protein function, a notion termed the classical structure-function paradigm. This book is devoted to demon-strating how this knowledge can be extended to understand how IDPs function. 1.1 PHYSICAL FORCES THAT SHAPE Protein Structure Because structural biology has its roots in studying globular (ordered) proteins, the classical concepts of Protein Structure are better suited for the description of ordered than disordered proteins. Usually, four hierarchical levels are distinguished, such as primary structure (sequence of amino acids in the polypeptide chain), secondary struc-ture (local, often repetitive structural elements [i.e., α -helix, β -strand, turn and coil]), tertiary structure (the fold in space of the entire polypeptide chain, also meaning the spatial arrangement of its secondary structural elements), and quaternary structure (stoichiometry and spatial arrangement of subunits in a multi-subunit protein).- Zdzislaw E. Sikorski(Author)
- 2001(Publication Date)
- CRC Press(Publisher)
The rela-tive spatial arrangement of amino acid residues is not taken into account at this level. The locations of any existing disulfide bridges should also be included here to complete a description of all covalent bonds, but in such a case, the IUPAC recommends a term “covalent structure.” The polypeptide chain is not free to take up any three-dimensional structure. Steric constraints and weak in-teractions, especially the formation of hydrogen bonds, determine that some spatial arrangements, i.e., conformations, are more stable than others. Secondary structure refers to regular recurring local conformations of adjacent amino acids in a polypeptide chain. There are a few common types of secondary structures, the most prominent being the a-helix and (3-conformation. Tertiary structure is the overall three-dimensional arrangement of the entire polypeptide chain. Usu-ally a few different types of secondary structures can be found within the ter-tiary structure of a large protein. Proteins with several polypeptide chains have one more level of structural organization: quaternary structure, which refers to the spatial relationship of the polypeptides, or subunits, within the protein. Continued advances in the understanding of Protein Structure and folding al-lowed two additional structural levels to be defined, both fall between sec-ondary and tertiary structural levels. A stable clustering of several elements of a secondary structure is called a supersecondary structure. These especially stable arrangements occur in many different proteins. A somewhat higher level of structural organization is the domain. This refers to a compact region that is a distinct structural and sometimes functional unit within a large polypep-tide chain. A monomeric protein may contain several domains that are often readily distinguishable within the overall structure. 3.4.2. THE PRIMARY AND SECONDARY STRUCTURES Proteins are composed of amino acids joined by amide (peptide) bonds.- eBook - PDF
- Lizabeth A. Allison(Author)
- 2021(Publication Date)
- Wiley-Blackwell(Publisher)
. . . . . O O Figure 4.9 Four levels of Protein Structure. The primary Protein Structure is the sequence of a chain of amino acids. Secondary structures such as the α -helix and the β -pleated sheet are stabilized by hydrogen bonding between nearby amino acids in the chain. The secondary structure folds into a three-dimensional tertiary structure through noncovalent and covalent interactions. The quaternary structure is a protein consisting of more than one amino acid chain. 4.3 The three-dimensional structure of proteins 91 the first enzyme ever to have its structure solved by X-ray diffraction. Lysozyme is a widespread enzyme found in animal secretions such as tears and in egg white. It catalyzes the breaking of glycosidic bonds between certain residues in components of bacterial cell walls, resulting in lysis of the bacteria. Because of its catalytic properties, lysozyme is C SH HS C C H 2 H 2 H 2 H 2 S S C Figure 4.10 Disulfide bonds in tertiary folding. The backbone structure of α -chymotrypsin, an enzyme involved in digesting proteins in the small intestine, is shown (Protein Data Bank, PDB: 5CHA). Its structure contains five disulfide bonds (red bars). Cysteines are shown in light orange. The inset shows two cysteine side chains on the opposite side of a loop domain. The two thiol groups can undergo a reaction involving the loss of two hydrogens and the formation of a covalent disulfide bond between them. Chymotrypsin is activated by cleavage of the inactive precursor chymotrypsinogen, which is secreted by the pancreas. The three segments of polypeptide chain (green, light blue, and dark blue) produced by proteolytic processing remain linked by disulfide bonds. Source: Based on Protein Data Bank, PDB: 5CHA. (a) Active site (b) (c) Figure 4.11 The structure of the globular protein lysozyme. (a) A ribbon model depicts how the α -helices (coiled ribbons) and β -pleated sheets (flat arrows) present in lysozyme interact to form a globular shape. - eBook - ePub
- David P. Clark(Author)
- 2009(Publication Date)
- Academic Cell(Publisher)
prosthetic groups —associated molecules that are not made of amino acids. The final shape of a protein is determined by its amino acid sequence, so proteins with similar sequences have similar 3-D conformations.Typical polypeptides are 300–400 amino acids long. Polypeptides much smaller or much larger are less common. However, many hormones and growth factors, such as insulin, do consist of relatively short polypeptide chains. Individual polypeptides with more than a thousand amino acids are very rare and very large proteins tend to consist of several separate polypeptide chains rather than a single long chain.Linear polypeptide chains are folded up to give the final 3D structures.The structures of biological polymers, both proteins and nucleic acids, are often divided into four levels of organization:1. Primary structure is the order of the monomers; i.e., the sequence of the amino acids for a protein, or of the nucleotides in the case of DNA or RNA.2. Secondary structure is the folding or coiling of the original polymer chains by means of hydrogen bonding. In the case of proteins, the hydrogen bonds are between the atoms of the polypeptide backbone.3. Tertiary structure is the further folding that gives the final 3-D structure of a single polymer chain. In the case of proteins, this involves interactions between the R groups of the amino acids.4. Quaternary structure is the assembly of several separate polymer chains.The Secondary Structure of Proteins Relies on Hydrogen Bonds
By definition, the secondary structure is folding that depends solely on hydrogen bonding. In DNA, hydrogen bonding occurs between base pairs and is the basis of the double helix. In proteins, hydrogen bonding occurs between the peptide groups that form the backbone of the polypeptide (Fig. 7.05 - eBook - PDF
- Clarence H. Suelter(Author)
- 2009(Publication Date)
- Wiley-Interscience(Publisher)
207 208 JOHN E. WILSON 1. INTRODUCTION The phrase “structure-function relationships” or its equivalent is frequently found in the literature of protein chemistry, and succinctly indicates what is a major objective for most investigators working in this area, namely, gaining an understanding of how specific structural features confer particular biological function. How is it that a certain array of amino acid residues (and in some cases, additional factors such as chelated metal ions or prosthetic groups) can harness basic thermodynamic forces to bring about a biologically relevant result that could never be accomplished on a meaningful time scale, if at all, in the absence of the protein? Aside from its intrinsic interest, an understanding of the molecular basis for such events is all the more important in a time when the techniques of protein engineering offer the potential for modifying structure, and thereby function, with a facility that would have seemed unthinkable only a few years ago. Obviously, the productive use of such methodology depends directly on a clear definition of the relationship between structure and function. The functions of proteins vary remarkably. Even if one were to consider only catalytically active proteins (enzymes), there is an incredible array of reactions that must occur to maintain the life of even the simplest organism - and an incredible array of organisms! If such diversity were also reflected in Protein Structure, with each enzyme possessing a structure totally unlike that of any other enzyme, it would seem a nearly hopeless task to ever develop any coherent set of principles governing the relationship between structure and function. - eBook - PDF
Biochemistry
An Integrative Approach
- John T. Tansey(Author)
- 2019(Publication Date)
- Wiley(Publisher)
In the lab- oratory, compounds such as β-mercaptoethanol (βME) or dithiothreitol (DTT) are often used to accomplish this task. In the cell, the cytosol is a reducing environment owing to the presence of the peptide glutathione. These compounds will reduce any disulfides that form. Proteins containing disul- fide bonds are synthesized and folded in the endoplasmic reticulum (ER). 3.3.4 The quaternary structure of a protein describes how individual subunits interact Most proteins exist in a complex, either with other subunits of a protein or with different proteins. This is termed the quaternary structure of the protein (Figure 3.24). The forces that govern the interactions of these individual subunits are the same as those involved in the formation of tertiary structure. FIGURE 3.24 Quaternary structure. The structure of purine nucleotide phosphorylases is an example of quaternary structure. This protein is a homotrimer with a3 stoichiometry. This enzyme has three identical subunits, each shown here in a different color. Note how the subunits are in close contact with each other in this example. (PDB ID 4EAR Haapalainen, A.M., Ho, M.C., Suarez, J.J., Almo, S.C., Schramm, V.L. (2013) Catalytic Site Conformations in Human PNP by (19) F-NMR and Crystallography. Chem.Biol. 20: 212–222) Three identical subunits (homotrimer, α3) 3.4 Examples of Protein Structures and Functions 89 3.3 Summary • Protein Structure can be described on four levels—primary through quaternary. • The basic unit of Protein Structure is the peptide bond. • Secondary structure describes higher order structures: a helices, β sheets, turns, and coils. • Secondary structures are stabilized by hydrogen bonds in the peptide backbone, and can be formed by many different amino acids. • Tertiary structures describe the interaction of different elements of helix and sheet to form a complex structure. • Tertiary structure represents the complete folding of a single polypeptide into a functional protein. - Available until 25 May |Learn more
Proteins
Concepts in Biochemistry
- Paulo Almeida(Author)
- 2016(Publication Date)
- Garland Science(Publisher)
2.4 THE SECONDARY STRUCTURE IS THE LOCAL SPATIAL ARRANGEMENT OF THE POLYPEPTIDE CHAIN In this section we begin to explore Protein Structure. Intuitively, most of us think of structure as arising from an organization of elements in space, the connections between those elements appearing from favorable, or attractive, interactions. We will see, however, that the concept of structure is much broader. Although favorable interactions do indeed occur in proteins, structure also arises from constraints . That is, by forbidding some conformations, others become the only possible ones for the polypeptide chain. The secondary structure of proteins is the result of restrictions on possible conformations and the requirement for hydrogen-bond formation. O C α C α N H : O C α C α N H Figure 2.30 The peptide group is a resonance hybrid of two chemical structures, which contribute unequally to the real structure. The Peptide Group is Planar The first restriction that we will encounter is on the peptide group itself. The peptide bond, between the nitrogen atom and the carbonyl carbon, is usually represented as a single bond (N −− C == O). This is misleading. Electrons are delocalized from the nitrogen atom to the carbonyl group by resonance, and the real structure is a hybrid of two resonance forms ( Figure 2.30 ). The two resonance forms do not con-tribute equally to the real structure. The form with an N −− C single bond 50 Chapter 2 Protein Structure contributes about 60%, whereas the form with an N == C double bond contributes about 40%. Therefore, the peptide bond has approximately 40% double-bond character. As a consequence of the partial double-bond character of the peptide bond, the two C α ’s, the N and its H, and the carbonyl C and its O are all in the same plane. The geometry of the peptide bond is shown in Figure 2.31 . The bond angles are close to 120 ◦ . The atoms inside the rectangle, which constitute the peptide group , are all in the same plane.
Index pages curate the most relevant extracts from our library of academic textbooks. They’ve been created using an in-house natural language model (NLM), each adding context and meaning to key research topics.